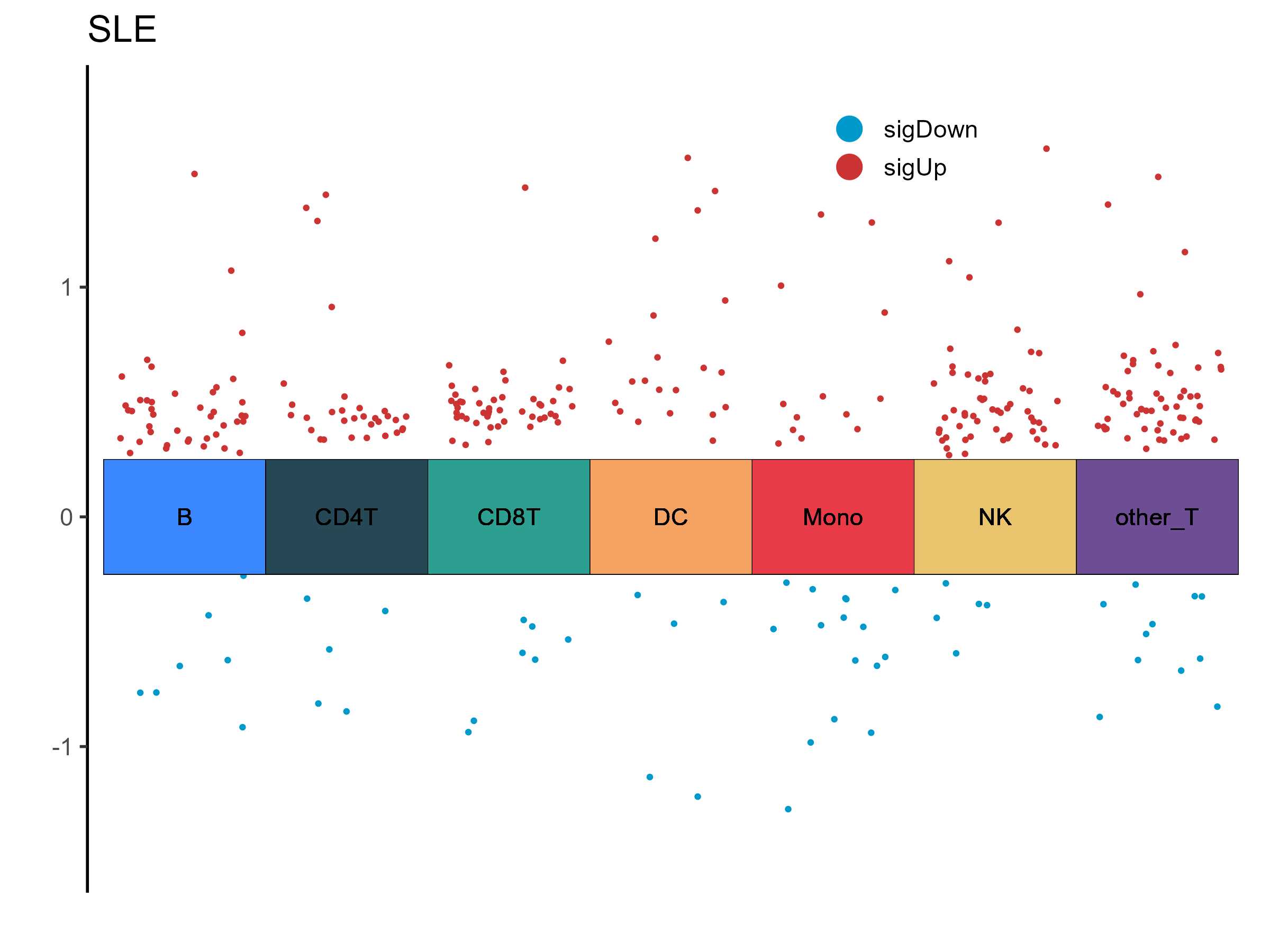
| B_8_2: Mitochondrial Diseases |
0.415344772316119 |
0.0437053757652906 |
B |
SLE |
|
| B_12_19: mitochondrial cytochrome c oxidase assembly |
0.369526773398977 |
0.0481936926885479 |
B |
SLE |
|
| B_12_14: Coatomer Protein |
0.306828533847793 |
0.0494469045437766 |
B |
SLE |
|
| B_14_27: NA |
-0.256237850611349 |
0.0495422021033727 |
B |
SLE |
|
| B_3_1: Protein processing in endoplasmic reticulum |
0.466866139872733 |
0.0522724743563323 |
B |
SLE |
|
| B_13_33: ATP metabolic process |
0.330292386029481 |
0.054117923133703 |
B |
SLE |
|
| B_14_45: immune response-activating cell surface receptor signaling pathway |
0.315966140680397 |
0.0544866988720069 |
B |
SLE |
|
| B_13_3: NA |
-0.290525204078106 |
0.0552207584249989 |
B |
SLE |
|
| B_14_26: DNA polymerase binding |
-0.255302639153515 |
0.0563449555595418 |
B |
SLE |
|
| B_6_1: Huntington disease |
0.406436687256688 |
0.056468319157812 |
B |
SLE |
|
| B_12_4: organelle disassembly |
0.262278886243883 |
0.0627744597489376 |
B |
SLE |
|
| B_13_36: CD4 Naive |
-0.254143881534682 |
0.0679954718868019 |
B |
SLE |
|
| B_14_7: heme biosynthetic process |
0.247316424480958 |
0.0693019149672034 |
B |
SLE |
|
| B_4_1: oxidative phosphorylation |
0.416521638483189 |
0.0724643210146857 |
B |
SLE |
|
| B_14_23: ras GTPase-Activating Proteins |
0.283966262333137 |
0.0738060153305841 |
B |
SLE |
|