SLE Volcano Plot
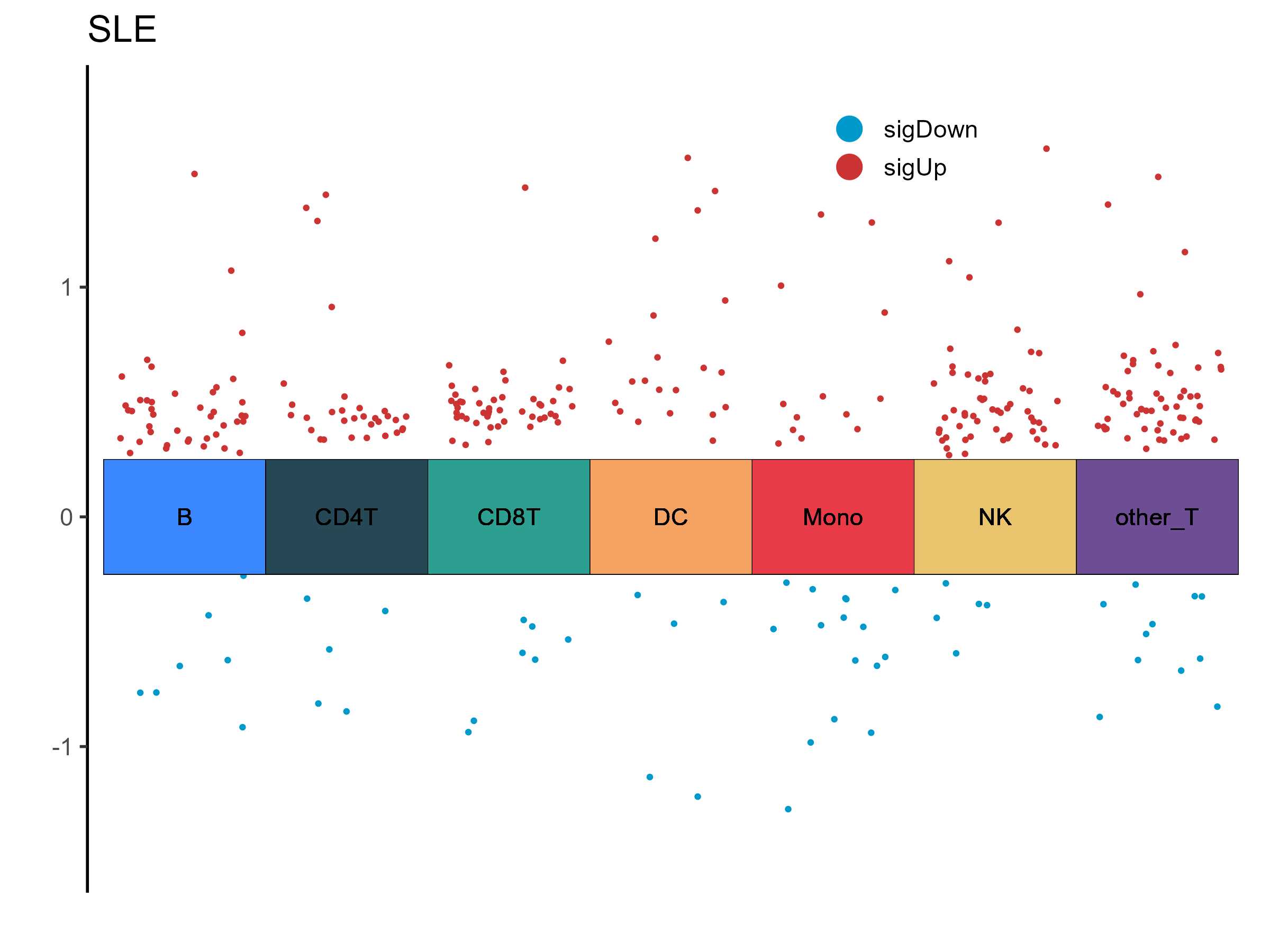
DEM Statistics (P < 0.05, |logFC| > 0.25)
| Cell Type | Up | Down | Total |
|---|---|---|---|
| B | 42 | 7 | 49 |
| other_T | 57 | 11 | 68 |
| Mono | 13 | 16 | 29 |
| NK | 56 | 5 | 61 |
| CD8T | 48 | 7 | 55 |
| DC | 21 | 5 | 26 |
| CD4T | 31 | 5 | 36 |
| Module_Name | logFC | P.Value | Cell_Type | disease | Expression |
|---|---|---|---|---|---|
| B_14_42: XBP1(S) activates chaperone genes | 0.361839960066934 | 0.11210791234616 | B | SLE | |
| B_14_1: TORC1 signaling | 0.180803534962737 | 0.116286765150269 | B | SLE | |
| B_13_24: NA | 0.221985291656653 | 0.123220611115774 | B | SLE | |
| B_13_22: Micronuclei, Chromosome-Defective | 0.245347750527847 | 0.149318962331699 | B | SLE | |
| B_13_10: Intercellular Junctions | 0.18153669479756 | 0.150842582376507 | B | SLE | |
| B_13_17: ubiquitin ligase complex | 0.174146428452402 | 0.151867182238322 | B | SLE | |
| B_13_35: Metabolic reprogramming in pancreatic cancer | 0.229452814357373 | 0.160038018705931 | B | SLE | |
| B_14_30: ribosome biogenesis | 0.205005892606881 | 0.162245354017323 | B | SLE | |
| B_14_15: regulation of natural killer cell proliferation | 0.213855799778365 | 0.170023068780859 | B | SLE | |
| B_12_7: RNA splicing | 0.330070232507832 | 0.175239445368112 | B | SLE | |
| B_5_1: Parkinson disease | 0.285589671216821 | 0.184497824014351 | B | SLE | |
| B_14_6: Receptors, Ectodysplasin | 0.177601167400668 | 0.19306436234835 | B | SLE | |
| B_14_24: DNA-templated DNA replication | 0.179346297131878 | 0.193185100497735 | B | SLE | |
| B_14_37: IL9 signaling | -0.257058754922368 | 0.209445916863176 | B | SLE | |
| B_13_7: MLL3/4 complex | 0.21452163135244 | 0.212283351189413 | B | SLE |