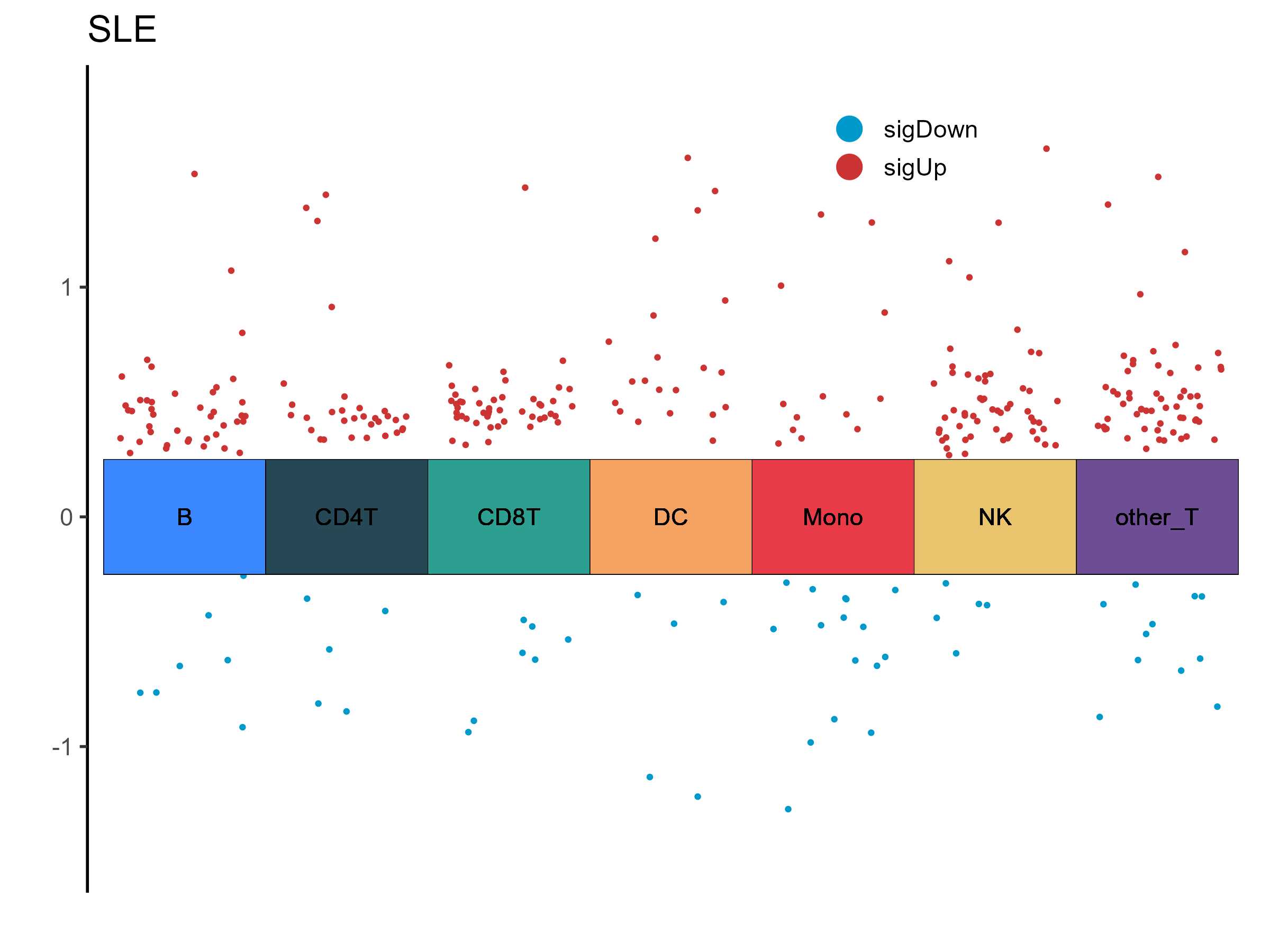
| B_14_11: negative regulation of double-strand break repair |
0.151015339318395 |
0.359656827356067 |
B |
SLE |
|
| B_11_12: Phosphoric Diester Hydrolases |
-0.163897499086042 |
0.363357820355998 |
B |
SLE |
|
| B_14_9: RNA Helicases |
0.137626296815014 |
0.394215126380429 |
B |
SLE |
|
| B_11_8: PAFAH1B1 copy number variation |
0.139718532864694 |
0.404665434658952 |
B |
SLE |
|
| B_13_15: cullin-RING ubiquitin ligase complex |
0.120107487207343 |
0.408909884734207 |
B |
SLE |
|
| B_12_9: Endocytosis |
0.118866654971906 |
0.426253005308429 |
B |
SLE |
|
| B_10_7: HALLMARK_E2F_TARGETS |
0.139793408476863 |
0.429287871810669 |
B |
SLE |
|
| B_13_18: mitochondrial translation |
0.145193372643434 |
0.433271207259038 |
B |
SLE |
|
| B_12_21: Organophosphates |
0.12090659103727 |
0.437136671288755 |
B |
SLE |
|
| B_10_1: RNA splicing |
0.137639275062271 |
0.445573451997442 |
B |
SLE |
|
| B_7_2: mitochondrial protein-containing complex |
0.161241431089954 |
0.452608184296955 |
B |
SLE |
|
| B_14_16: protein-folding chaperone binding |
0.0962510288851544 |
0.480323664665788 |
B |
SLE |
|
| B_9_1: alternative mRNA splicing, via spliceosome |
0.131508986802668 |
0.486293795694651 |
B |
SLE |
|
| B_13_34: protein sequestering activity |
0.157005857868208 |
0.495168371594522 |
B |
SLE |
|
| B_10_6: Signaling by TGFBR3 |
-0.113104110961822 |
0.508691246388432 |
B |
SLE |
|