SLE Volcano Plot
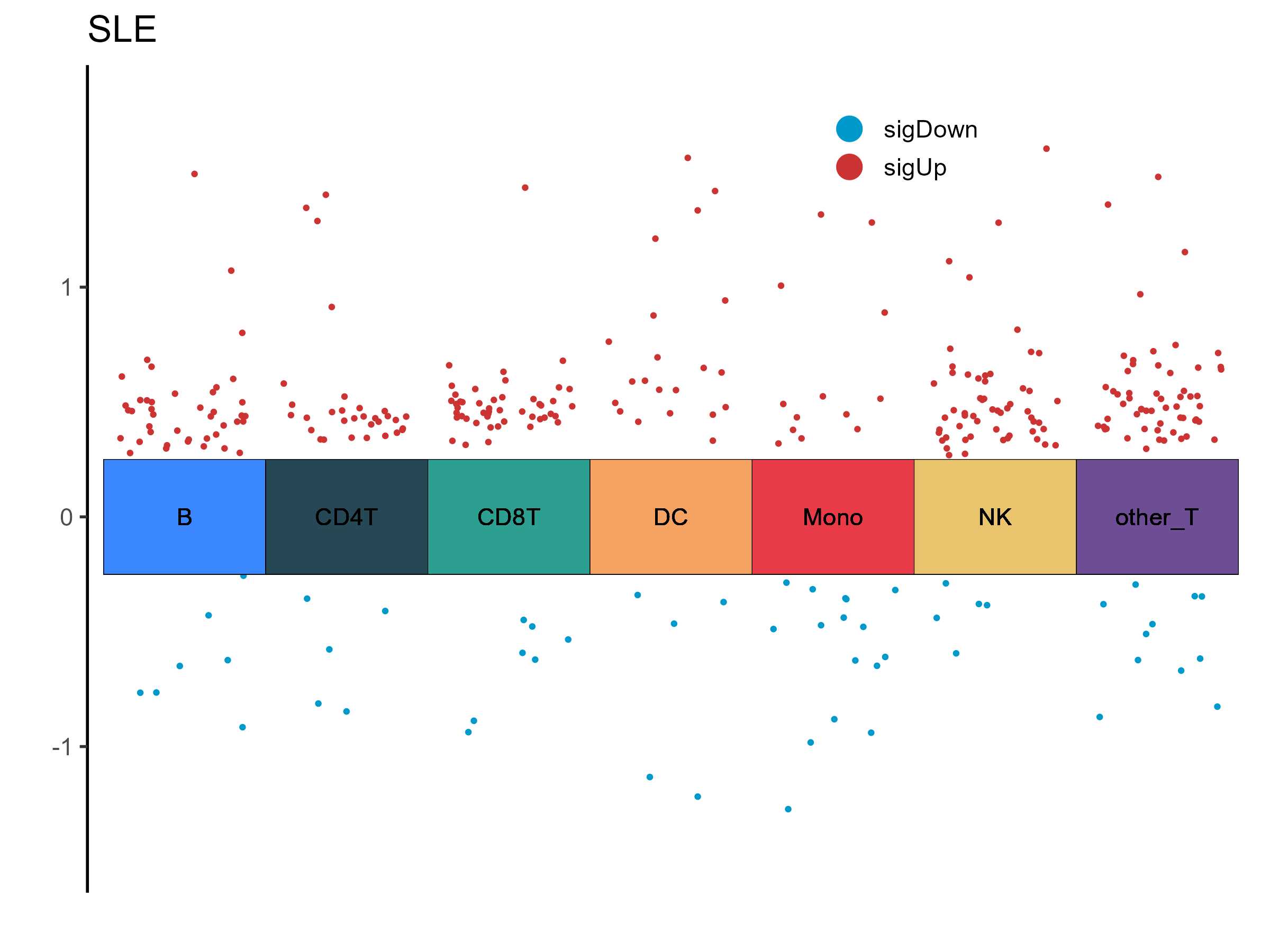
DEM Statistics (P < 0.05, |logFC| > 0.25)
| Cell Type | Up | Down | Total |
|---|---|---|---|
| B | 42 | 7 | 49 |
| other_T | 57 | 11 | 68 |
| Mono | 13 | 16 | 29 |
| NK | 56 | 5 | 61 |
| CD8T | 48 | 7 | 55 |
| DC | 21 | 5 | 26 |
| CD4T | 31 | 5 | 36 |
| Module_Name | logFC | P.Value | Cell_Type | disease | Expression |
|---|---|---|---|---|---|
| B_13_25: NA | 0.18400268625285 | 0.213875169190092 | B | SLE | |
| B_13_30: core mediator complex | 0.209166065140627 | 0.214590195056903 | B | SLE | |
| B_13_6: B memory | 0.19626550137083 | 0.250622073757109 | B | SLE | |
| B_11_14: ARF protein signal transduction | 0.15984569468913 | 0.261514240555023 | B | SLE | |
| B_14_25: Dictyostelium | 0.191142558083669 | 0.265943919743034 | B | SLE | |
| B_13_32: Genes related to primary cilium development based on CRISPR | 0.160707353483032 | 0.266864781203128 | B | SLE | |
| B_14_34: NK | 0.150360181254343 | 0.277512100221359 | B | SLE | |
| B_14_31: HALLMARK_DNA_REPAIR | 0.192394316187771 | 0.303829655137403 | B | SLE | |
| B_13_37: ubiquitin conjugating enzyme activity | 0.179952816134691 | 0.318127150968036 | B | SLE | |
| B_13_13: Immunologic Deficiency Syndromes | -0.143854649196242 | 0.323672524651364 | B | SLE | |
| B_11_7: NK | -0.187951078867275 | 0.334252030629203 | B | SLE | |
| B_11_9: Termination of translesion DNA synthesis | 0.182606121243011 | 0.335624427818474 | B | SLE | |
| B_13_2: GATOR2 complex | -0.155739952052453 | 0.3462084217572 | B | SLE | |
| B_9_3: mitochondrial protein-containing complex | 0.188591691317833 | 0.348541941480459 | B | SLE | |
| B_11_3: B naive | -0.158134672139037 | 0.350095696043068 | B | SLE |