Module Explorer
Instructions
- Enter a module name.
- Click Submit to search for the detailed information of the module.
- Results will appear below if available.
Selected Module: B_13_37
Module Info
| Module_Name | B_13_37 |
| Cell_Type | B |
| Name | B_13_37: ubiquitin conjugating enzyme activity |
| Meta_Module | B_3 |
| Gene_Number | 49 |
Genes:
ACTN4, ARL6IP4, ASL, ATP5F1E, ATP6V1F, BRI3BP, CALM1, CAPNS1, CLIC1, CLN6, COPS4, CPNE5, CRLS1, CXXC1, DECR1, DGCR6L, DOK3, EIF3K, EIPR1, FAM50A, H3-3A, HDGF, HSPBP1, JPT1, KPNA2, LSM1, MACROH2A1, MRPS24, MT1X, MZT2A, PCK2, RAC1, S100A10, SH2B2, SMC4, SMIM12, SSU72, TCEAL8, THOC3, THYN1, TMA7, TMEM223, TRPV2, TSG101, TXNDC9, UBE2A, UBE2E3, UBE2J2, UBE2S
UMAP Visualizations

Word Cloud
Enriched TF
No TF enriched
Functional Enrichment Results
|
enrichment_term
|
module | ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count |
|---|---|---|---|---|---|---|---|---|---|---|
| GO | B_13_37 | GO:0061631 | ubiquitin conjugating enzyme activity | 4/48 | 34/18737 | 1.66130404775016e-06 | 0.00025249553885104 | 0.00020956107475491 | UBE2A/UBE2E3/UBE2J2/UBE2S | 4 |
| GO | B_13_37 | GO:0061650 | ubiquitin-like protein conjugating enzyme activity | 4/48 | 40/18737 | 3.23712229296217e-06 | 0.00025249553885104 | 0.00020956107475491 | UBE2A/UBE2E3/UBE2J2/UBE2S | 4 |
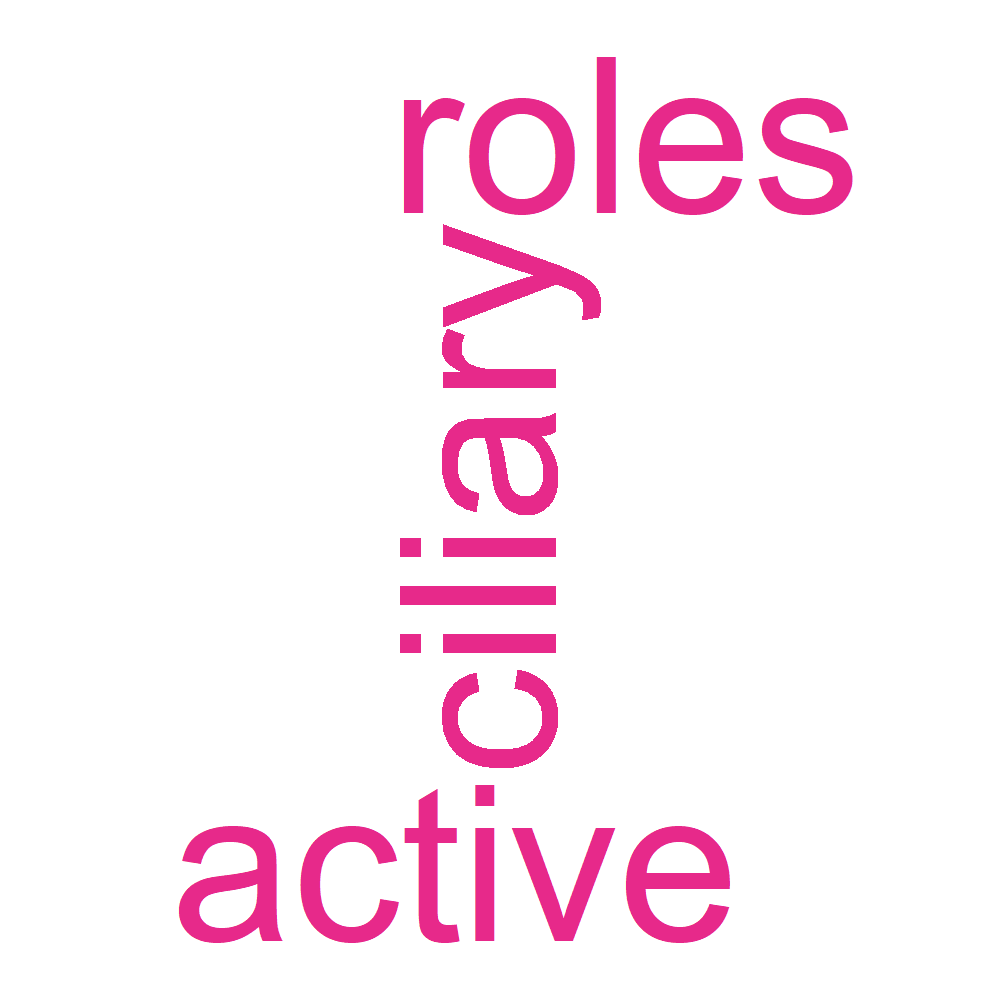
| WikiPathways | B_13_37 | WP4352 | Ciliary landscape | 6/30 | 216/8836 | 7.23317025607708e-05 | 0.0107050919789941 | 0.00966960655286094 | CALM1/COPS4/EIPR1/H3-3A/RAC1/SMC4 | 6 |
| reactome | B_13_37 | R-HSA-8852135 | Protein ubiquitination | 4/34 | 79/11146 | 9.22325640514363e-05 | 0.0185127757367803 | 0.0155582750249682 | UBE2A/UBE2E3/UBE2J2/UBE2S | 4 |
| reactome | B_13_37 | R-HSA-8866652 | Synthesis of active ubiquitin: roles of E1 and E2 enzymes | 3/34 | 30/11146 | 9.95310523482813e-05 | 0.0185127757367803 | 0.0155582750249682 | UBE2A/UBE2E3/UBE2S | 3 |
| reactome | B_13_37 | R-HSA-442729 | CREB1 phosphorylation through the activation of CaMKII/CaMKK/CaMKIV cascasde | 2/34 | 10/11146 | 0.00040026981428013 | 0.0496334569707365 | 0.041712328014456 | CALM1/KPNA2 | 2 |
1