SLE Volcano Plot
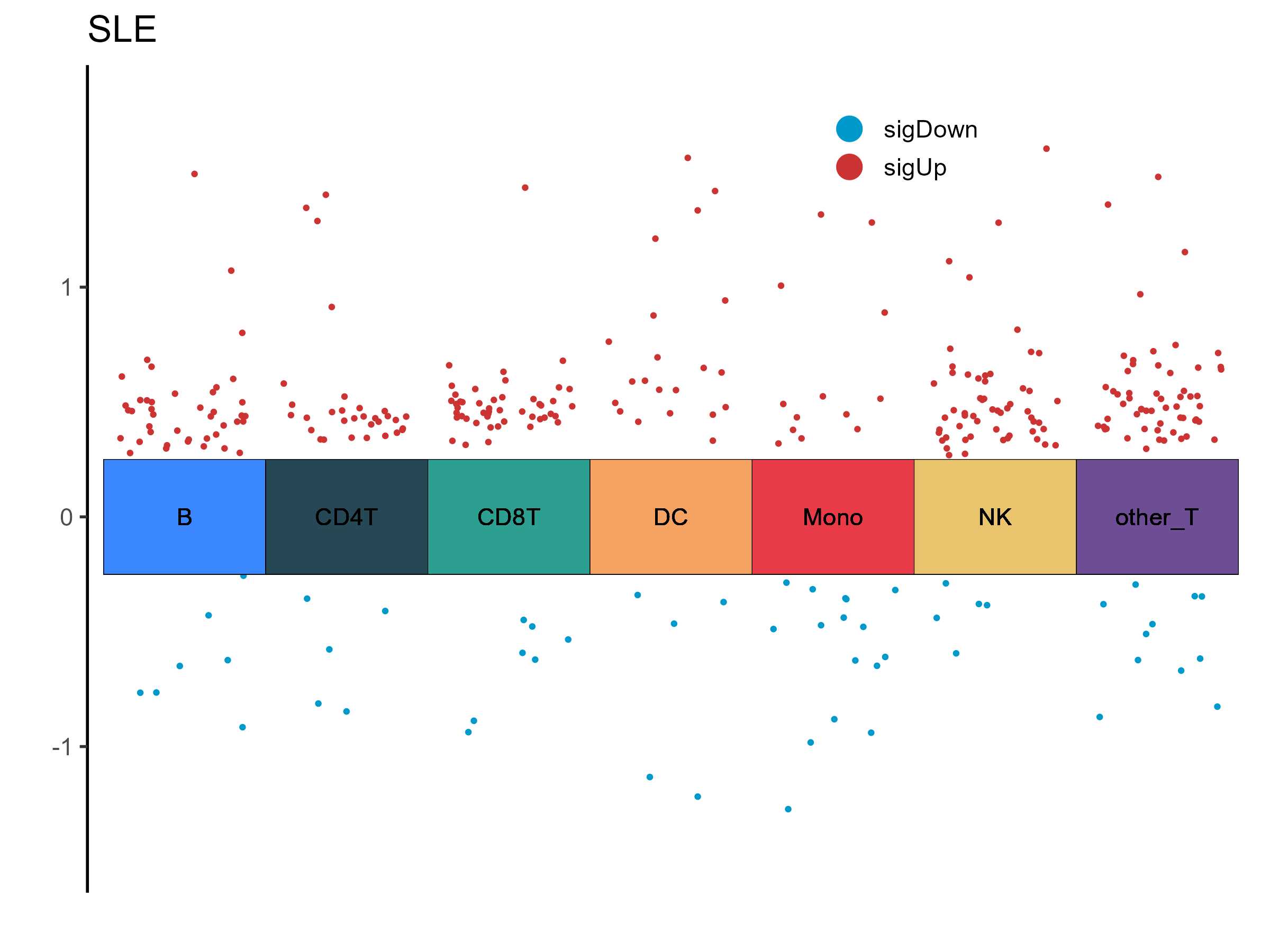
DEM Statistics (P < 0.05, |logFC| > 0.25)
| Cell Type | Up | Down | Total |
|---|---|---|---|
| B | 42 | 7 | 49 |
| other_T | 57 | 11 | 68 |
| Mono | 13 | 16 | 29 |
| NK | 56 | 5 | 61 |
| CD8T | 48 | 7 | 55 |
| DC | 21 | 5 | 26 |
| CD4T | 31 | 5 | 36 |
| Module_Name | logFC | P.Value | Cell_Type | disease | Expression |
|---|---|---|---|---|---|
| B_12_15: response to virus | 1.49291579546123 | 6.73402894907904e-09 | B | SLE | |
| B_10_8: Calgranulin B | 1.07193148291602 | 9.54174661531841e-07 | B | SLE | |
| B_14_21: Immunologic Factors | 0.801041489874594 | 2.61988620044957e-06 | B | SLE | |
| B_11_10: Golgi stack | 0.610963103043583 | 6.25034942469471e-05 | B | SLE | |
| B_13_1: regulation of ribonucleoprotein complex localization | 0.600627186060267 | 0.00013213168411984 | B | SLE | |
| B_10_11: Eukaryotic Initiation Factors | -0.765621488772213 | 0.000152407211990279 | B | SLE | |
| B_11_11: B intermediate | 0.653698640265052 | 0.000667708100640958 | B | SLE | |
| B_14_13: Bcl-2 family protein complex | 0.499607538823037 | 0.000834514080726703 | B | SLE | |
| B_1_1: RNA, Nuclear | -0.915755706092987 | 0.00151062991738075 | B | SLE | |
| B_6_2: Asparagine N-linked glycosylation | 0.683618082598411 | 0.00194082163415765 | B | SLE | |
| B_13_23: Endosomal/Vacuolar pathway | 0.440857395245585 | 0.00265438356284645 | B | SLE | |
| B_14_40: HALLMARK_HEME_METABOLISM | 0.456636732379691 | 0.00283403198312567 | B | SLE | |
| B_13_4: Muridae | 0.445608865290759 | 0.00372594000241268 | B | SLE | |
| B_3_2: cytoplasmic translation | -0.764679158378527 | 0.00385702775997167 | B | SLE | |
| B_7_1: B naive | -0.649303458703091 | 0.00402891910887948 | B | SLE |