SLE Volcano Plot
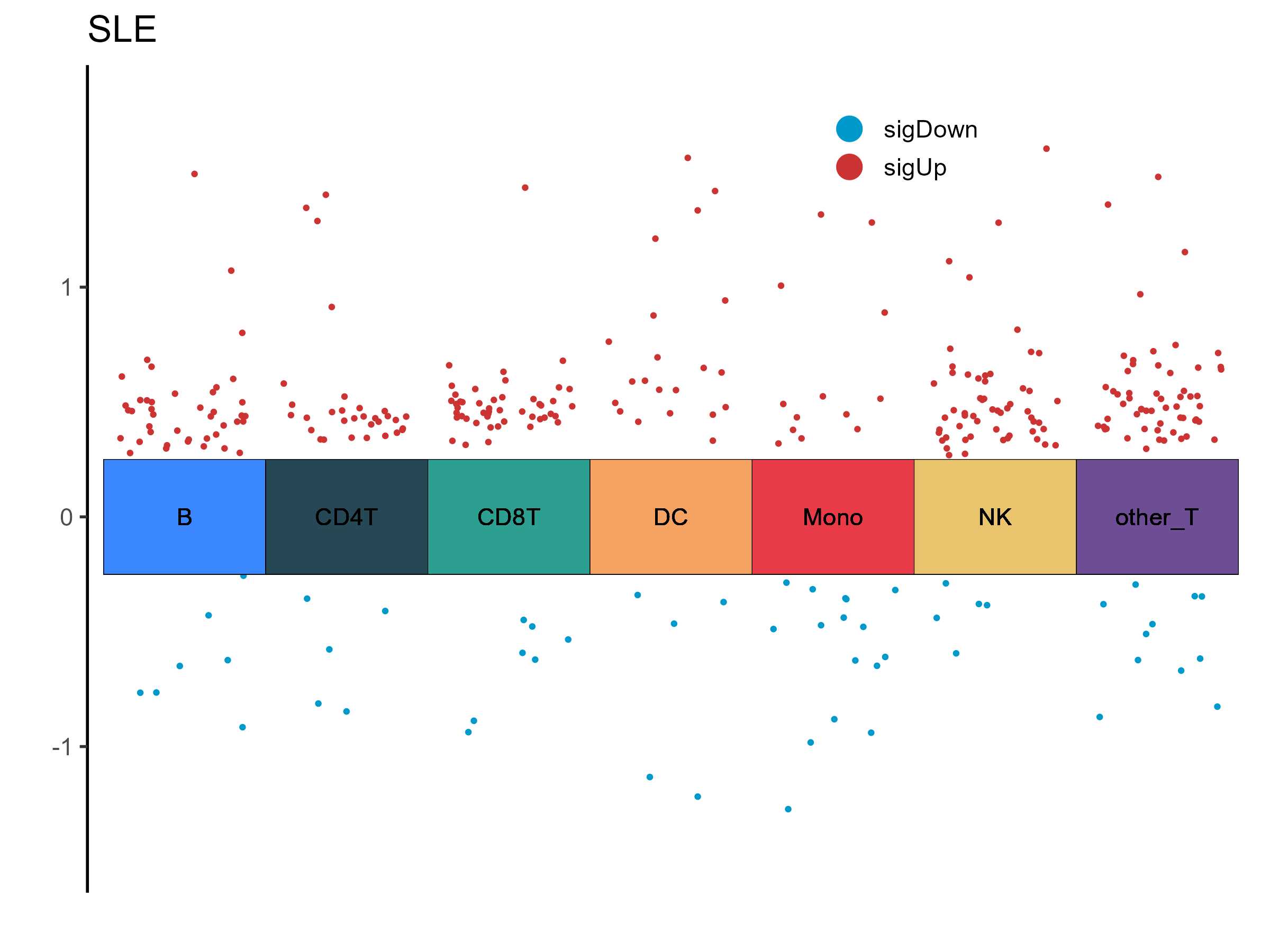
DEM Statistics (P < 0.05, |logFC| > 0.25)
| Cell Type | Up | Down | Total |
|---|---|---|---|
| B | 42 | 7 | 49 |
| other_T | 57 | 11 | 68 |
| Mono | 13 | 16 | 29 |
| NK | 56 | 5 | 61 |
| CD8T | 48 | 7 | 55 |
| DC | 21 | 5 | 26 |
| CD4T | 31 | 5 | 36 |
| Module_Name | logFC | P.Value | Cell_Type | disease | Expression |
|---|---|---|---|---|---|
| B_14_46: Platelet activation, signaling and aggregation | 0.0698008583971564 | 0.721862986264826 | B | SLE | |
| B_12_2: Genes, Insect | -0.0492052918039918 | 0.723625660760168 | B | SLE | |
| B_14_38: Situs Inversus | -0.0472994635503948 | 0.743811251955907 | B | SLE | |
| B_12_16: p53 transcriptional gene network | 0.049071384971028 | 0.750846945315433 | B | SLE | |
| B_14_29: HALLMARK_KRAS_SIGNALING_UP | 0.0404656563412753 | 0.753409553426521 | B | SLE | |
| B_11_1: CCR4-NOT complex | -0.0710456518707008 | 0.75913893032589 | B | SLE | |
| B_14_18: ribosome biogenesis | 0.0333346803115324 | 0.798020003341314 | B | SLE | |
| B_10_10: HALLMARK_TNFA_SIGNALING_VIA_NFKB | 0.0612858893481772 | 0.823538593928206 | B | SLE | |
| B_12_17: NA | 0.0317856999439423 | 0.835010630917865 | B | SLE | |
| B_13_29: NA | 0.0279240494829869 | 0.844877536289416 | B | SLE | |
| B_13_21: positive regulation of NLRP3 inflammasome complex assembly | 0.0311659797583792 | 0.853856540599431 | B | SLE | |
| B_13_31: alpha-beta T cell differentiation | -0.0362726674367734 | 0.860674531928099 | B | SLE | |
| B_14_12: nucleocytoplasmic transport | 0.021837197135412 | 0.869824627833715 | B | SLE | |
| B_7_5: protein-DNA complex organization | 0.0321705562550254 | 0.87844599213093 | B | SLE | |
| B_12_20: N-acetyltransferase activity | 0.017745680603224 | 0.922115465109234 | B | SLE |