Module Explorer
Instructions
- Enter a module name.
- Click Submit to search for the detailed information of the module.
- Results will appear below if available.
Selected Module: B_9_3
Module Info
| Module_Name | B_9_3 |
| Cell_Type | B |
| Name | B_9_3: mitochondrial protein-containing complex |
| Meta_Module | B_3 |
| Gene_Number | 67 |
Genes:
ADSL, ALDOA, ATP1B3, CCND2, CCT5, CCT7, CDK4, CHCHD7, COA6, EBNA1BP2, EEF1E1, EIF3I, EIF5A, ENY2, FABP5, GADD45GIP1, GGCT, GNL2, GRWD1, HADH, HIGD1A, HSPD1, ILF2, IMP4, LDHA, LSM5, MAGOHB, MICOS10, MRPL12, MRPL16, MRPL17, MRPL18, MRPL4, MRPS25, NDUFAF8, NEDD8, NOL7, OXCT1, PAFAH1B3, PAICS, PDAP1, PDCL3, PGK1, PHB1, PKM, POLR2F, POLR2H, POLR2I, POLR2J, PPM1G, PRMT1, PSMA7, PSMF1, RBIS, RPA3, TAF9, TIMM10, TIMM13, TIMMDC1, TIPRL, TOMM22, TOMM40, TRAP1, UBA1, UQCRFS1, VBP1, VDAC2
UMAP Visualizations

Word Cloud
Enriched TF
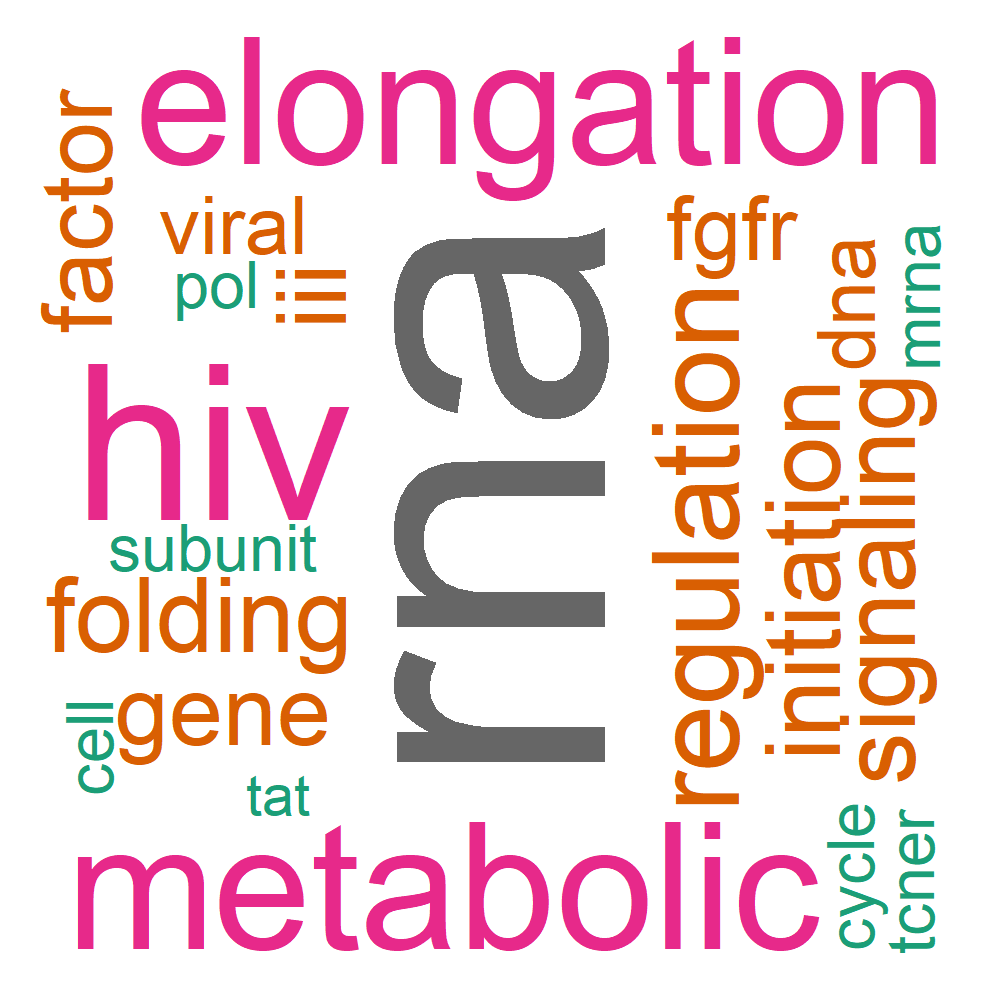
| TF | p.adjust | Genes |
|---|---|---|
| TGACCTTG | 0.00077214560723105 | ALDOA/HIGD1A/HSPD1/TOMM40/TRAP1/UBA1/VDAC2 |
| EPC1 | 0.00077214560723105 | COA6/EIF5A/MRPL4/PDAP1/PKM/POLR2J/PRMT1/TIMM13 |
| USF | 0.00077214560723105 | EEF1E1/EIF5A/PAICS/POLR2H/PRMT1/TIMM10/UBA1 |
| ATF5 | 0.00162546136866258 | ALDOA/CCT5/HSPD1/ILF2/LSM5/MRPL18/RBIS/RPA3 |
| HIF1 | 0.00190031811966115 | ATP1B3/CCND2/LDHA/PGK1/PRMT1/UBA1 |
| HSF2 | 0.00209156428902918 | CCT5/CCT7/HSPD1/MRPL18/PGK1/PHB1 |
| MYBL1 | 0.00949528706847933 | ALDOA/CCND2/CDK4/IMP4/PKM/RPA3/TOMM40 |
| CASP8AP2 | 0.00949528706847933 | COA6/HSPD1/ILF2/LSM5/MRPL16/PSMF1 |
Functional Enrichment Results
|
enrichment_term
|
module | ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count |
|---|---|---|---|---|---|---|---|---|---|---|
| reactome | B_9_3 | R-HSA-168325 | Viral Messenger RNA Synthesis | 4/60 | 43/11146 | 8.00384904279607e-05 | 0.00085374389789824 | 0.00055951333389586 | POLR2F/POLR2H/POLR2I/POLR2J | 4 |
| reactome | B_9_3 | R-HSA-5655253 | Signaling by FGFR2 in disease | 4/60 | 43/11146 | 8.00384904279607e-05 | 0.00085374389789824 | 0.00055951333389586 | POLR2F/POLR2H/POLR2I/POLR2J | 4 |
| MeSH_gene2pubmed | B_9_3 | D033681 | Mitochondrial Membrane Transport Proteins | 4/67 | 90/26076 | 8.61300928243825e-05 | 0.00294860546184548 | 0.00246863931105898 | TIMM10/TIMM13/TOMM22/VDAC2 | 4 |
| MeSH_gene2pubmed | B_9_3 | D013539 | Simian virus 40 | 7/67 | 410/26076 | 8.71575056934773e-05 | 0.00294860546184548 | 0.00246863931105898 | CCND2/CDK4/POLR2F/POLR2H/POLR2I/POLR2J/TAF9 | 7 |
| MeSH_gene2pubmed | B_9_3 | D000260 | Adenoviruses, Human | 6/67 | 284/26076 | 9.05104552860466e-05 | 0.00301564380566692 | 0.0025247653316634 | CDK4/POLR2F/POLR2H/POLR2I/POLR2J/TAF9 | 6 |
| hallmark | B_9_3 | HALLMARK_MYC_TARGETS_V2 | HALLMARK_MYC_TARGETS_V2 | 5/36 | 58/4384 | 9.37459308521877e-05 | 0.00107807820480016 | 0.00083877938130904 | CDK4/GRWD1/HSPD1/IMP4/PHB1 | 5 |
| reactome | B_9_3 | R-HSA-167152 | Formation of HIV elongation complex in the absence of HIV Tat | 4/60 | 45/11146 | 9.5859295802484e-05 | 0.00099693667634583 | 0.00065335677928535 | POLR2F/POLR2H/POLR2I/POLR2J | 4 |
| MeSH_gene2pubmed | B_9_3 | D003673 | Defective Viruses | 4/67 | 93/26076 | 9.78509913852956e-05 | 0.00321155716501888 | 0.00268878843570907 | POLR2F/POLR2H/POLR2I/POLR2J | 4 |
| GO | B_9_3 | GO:0070585 | protein localization to mitochondrion | 5/66 | 133/18986 | 9.92489837189272e-05 | 0.00647245158109861 | 0.00523855538125465 | HSPD1/TIMM10/TIMM13/TOMM22/TOMM40 | 5 |
| GO | B_9_3 | GO:0009988 | cell-cell recognition | 4/66 | 73/18986 | 0.00012104764315499 | 0.00736776654670059 | 0.0059631891575302 | ALDOA/CCT5/CCT7/VDAC2 | 4 |