Module Explorer
Instructions
- Enter a module name.
- Click Submit to search for the detailed information of the module.
- Results will appear below if available.
Selected Module: B_9_3
Module Info
| Module_Name | B_9_3 |
| Cell_Type | B |
| Name | B_9_3: mitochondrial protein-containing complex |
| Meta_Module | B_3 |
| Gene_Number | 67 |
Genes:
ADSL, ALDOA, ATP1B3, CCND2, CCT5, CCT7, CDK4, CHCHD7, COA6, EBNA1BP2, EEF1E1, EIF3I, EIF5A, ENY2, FABP5, GADD45GIP1, GGCT, GNL2, GRWD1, HADH, HIGD1A, HSPD1, ILF2, IMP4, LDHA, LSM5, MAGOHB, MICOS10, MRPL12, MRPL16, MRPL17, MRPL18, MRPL4, MRPS25, NDUFAF8, NEDD8, NOL7, OXCT1, PAFAH1B3, PAICS, PDAP1, PDCL3, PGK1, PHB1, PKM, POLR2F, POLR2H, POLR2I, POLR2J, PPM1G, PRMT1, PSMA7, PSMF1, RBIS, RPA3, TAF9, TIMM10, TIMM13, TIMMDC1, TIPRL, TOMM22, TOMM40, TRAP1, UBA1, UQCRFS1, VBP1, VDAC2
UMAP Visualizations

Word Cloud
Enriched TF
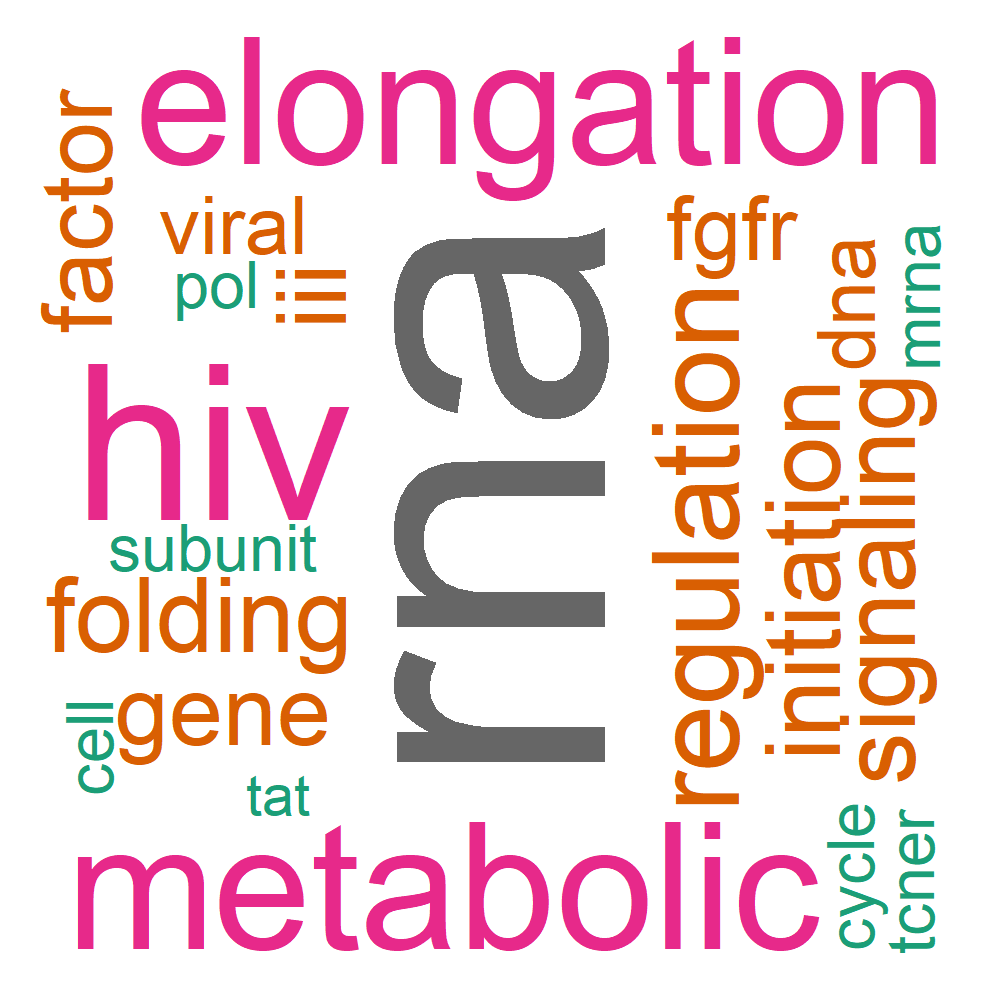
| TF | p.adjust | Genes |
|---|---|---|
| TGACCTTG | 0.00077214560723105 | ALDOA/HIGD1A/HSPD1/TOMM40/TRAP1/UBA1/VDAC2 |
| EPC1 | 0.00077214560723105 | COA6/EIF5A/MRPL4/PDAP1/PKM/POLR2J/PRMT1/TIMM13 |
| USF | 0.00077214560723105 | EEF1E1/EIF5A/PAICS/POLR2H/PRMT1/TIMM10/UBA1 |
| ATF5 | 0.00162546136866258 | ALDOA/CCT5/HSPD1/ILF2/LSM5/MRPL18/RBIS/RPA3 |
| HIF1 | 0.00190031811966115 | ATP1B3/CCND2/LDHA/PGK1/PRMT1/UBA1 |
| HSF2 | 0.00209156428902918 | CCT5/CCT7/HSPD1/MRPL18/PGK1/PHB1 |
| MYBL1 | 0.00949528706847933 | ALDOA/CCND2/CDK4/IMP4/PKM/RPA3/TOMM40 |
| CASP8AP2 | 0.00949528706847933 | COA6/HSPD1/ILF2/LSM5/MRPL16/PSMF1 |
Functional Enrichment Results
|
enrichment_term
|
module | ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count |
|---|---|---|---|---|---|---|---|---|---|---|
| reactome | B_9_3 | R-HSA-6807505 | RNA polymerase II transcribes snRNA genes | 5/60 | 74/11146 | 4.62384285983791e-05 | 0.00060109957177892 | 0.00039393924365066 | POLR2F/POLR2H/POLR2I/POLR2J/TAF9 | 5 |
| MeSH_gene2pubmed | B_9_3 | D002628 | Chemoreceptor Cells | 4/67 | 77/26076 | 4.67733733865847e-05 | 0.00177335600132931 | 0.00148469383036546 | POLR2F/POLR2H/POLR2I/POLR2J | 4 |
| GO | B_9_3 | GO:0006091 | generation of precursor metabolites and energy | 9/66 | 490/18986 | 4.73188831129146e-05 | 0.00360017835684092 | 0.00291384701274264 | ADSL/ALDOA/COA6/LDHA/OXCT1/PGK1/PKM/TRAP1/UQCRFS1 | 9 |
| WikiPathways | B_9_3 | WP534 | Glycolysis and gluconeogenesis | 4/41 | 45/8836 | 5.18103744950852e-05 | 0.00101642472969991 | 0.00080445170534223 | ALDOA/LDHA/PGK1/PKM | 4 |
| GO | B_9_3 | GO:0005742 | mitochondrial outer membrane translocase complex | 3/67 | 22/19960 | 5.31848332615401e-05 | 0.00051307721499368 | 0.000312851960362 | MICOS10/TOMM22/TOMM40 | 3 |
| reactome | B_9_3 | R-HSA-6781827 | Transcription-Coupled Nucleotide Excision Repair (TC-NER) | 5/60 | 77/11146 | 5.60177948341502e-05 | 0.00070616371669716 | 0.00046279454105406 | POLR2F/POLR2H/POLR2I/POLR2J/RPA3 | 5 |
| MeSH_gene2pubmed | B_9_3 | D009492 | Neurospora crassa | 4/67 | 81/26076 | 5.70623070118323e-05 | 0.00212677988337321 | 0.00178058831335762 | EIF5A/TIMM13/TOMM22/VDAC2 | 4 |
| MeSH_gene2pubmed | B_9_3 | D003676 | Deferoxamine | 5/67 | 161/26076 | 5.97861019001376e-05 | 0.00219116063464004 | 0.00183448933725159 | CDK4/EIF5A/LDHA/TRAP1/VBP1 | 5 |
| WikiPathways | B_9_3 | WP1946 | Cori cycle | 3/41 | 17/8836 | 6.02787735123976e-05 | 0.00101642472969991 | 0.00080445170534223 | ALDOA/LDHA/PGK1 | 3 |
| GO | B_9_3 | GO:0098799 | outer mitochondrial membrane protein complex | 3/67 | 23/19960 | 6.10157920183068e-05 | 0.00055592166061124 | 0.00033897662232392 | MICOS10/TOMM22/TOMM40 | 3 |