Module Explorer
Instructions
- Enter a module name.
- Click Submit to search for the detailed information of the module.
- Results will appear below if available.
Selected Module: B_8_2
Module Info
| Module_Name | B_8_2 |
| Cell_Type | B |
| Name | B_8_2: Mitochondrial Diseases |
| Meta_Module | B_3 |
| Gene_Number | 48 |
Genes:
ABRACL, ANAPC5, AP1S1, C1orf43, CBR1, COMMD4, DDX1, DDX49, DPM1, EIF4E2, FH, FUNDC2, IDH3A, MRPL14, MRPL28, MRPL3, MRPL32, MRPL58, MRPS18A, NABP2, NDUFA11, NDUFA8, NDUFAF3, NDUFB9, NDUFS3, NDUFS7, NDUFV1, PFDN6, POLR2E, PPP1R7, PSMA1, PSMB5, PSMC1, PSMD9, RNH1, RUVBL2, SKA2, SLC25A39, SLX9, SRPK1, STARD7, TALDO1, TIMM44, TPGS2, TPI1, USP14, VPS25, ZCRB1
UMAP Visualizations

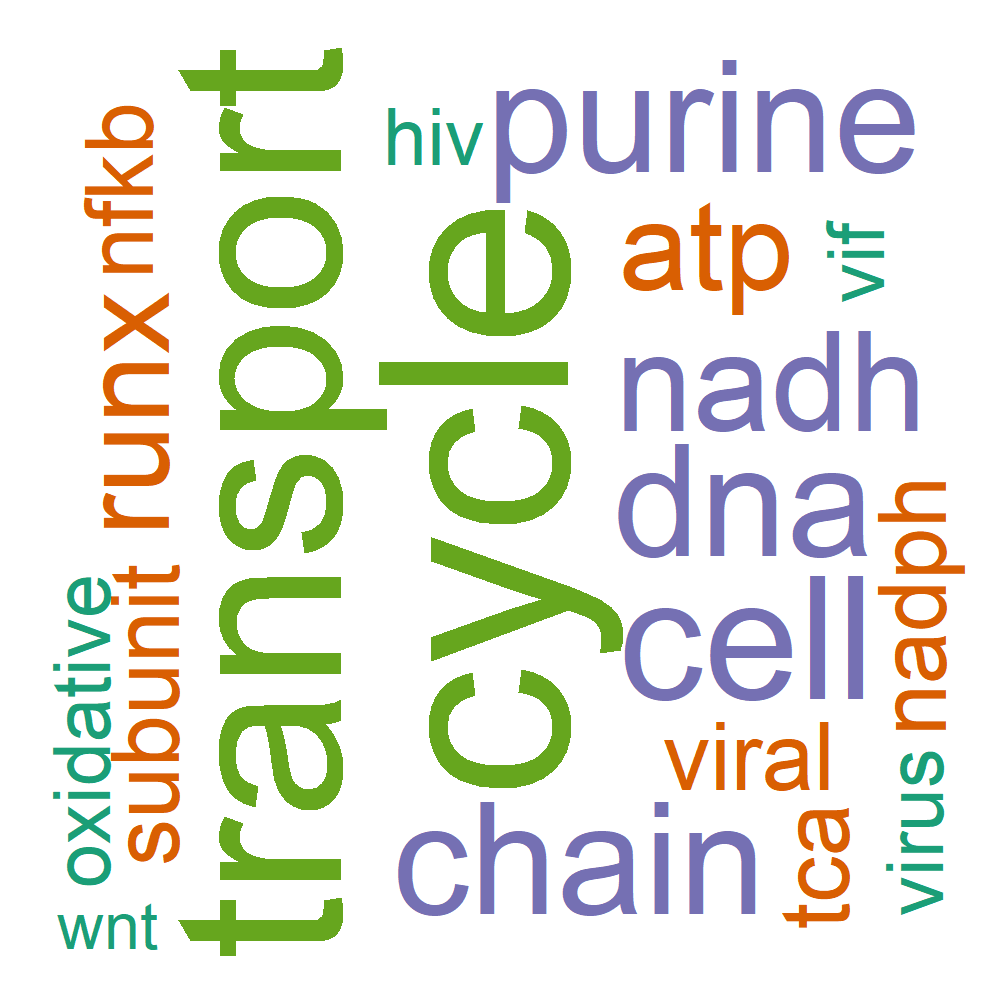
Word Cloud
Enriched TF
| TF | p.adjust | Genes |
|---|---|---|
| RLF | 4.65826687298153e-05 | ANAPC5/EIF4E2/MRPL3/MRPL58/NDUFS3/NDUFS7/PSMA1/SLX9 |
| ALKBH3 | 0.00163190543184927 | DDX1/EIF4E2/MRPL3/MRPL58/NDUFS7/SLX9/VPS25 |
| ZNF85 | 0.0251523044088028 | ABRACL/NABP2/NDUFS3/NDUFS7/VPS25 |
Functional Enrichment Results
|
enrichment_term
|
module | ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count |
|---|---|---|---|---|---|---|---|---|---|---|
| reactome | B_8_2 | R-HSA-5368287 | Mitochondrial translation | 6/41 | 93/11146 | 1.01901436116318e-06 | 5.57061184102541e-05 | 3.2715724226818e-05 | MRPL14/MRPL28/MRPL3/MRPL32/MRPL58/MRPS18A | 6 |
| GO | B_8_2 | GO:0046034 | ATP metabolic process | 7/47 | 228/18986 | 1.37312399814427e-06 | 5.20171679297007e-05 | 4.29366946788147e-05 | NDUFA11/NDUFA8/NDUFB9/NDUFS3/NDUFS7/NDUFV1/TPI1 | 7 |
| reactome | B_8_2 | R-HSA-9837999 | Mitochondrial protein degradation | 6/41 | 98/11146 | 1.38832150245198e-06 | 6.19179689650304e-05 | 3.63638906309389e-05 | FH/IDH3A/MRPL32/NDUFS3/NDUFV1/STARD7 | 6 |
| reactome | B_8_2 | R-HSA-611105 | Respiratory electron transport | 7/41 | 158/11146 | 1.51019436500074e-06 | 6.19179689650304e-05 | 3.63638906309389e-05 | NDUFA11/NDUFA8/NDUFAF3/NDUFB9/NDUFS3/NDUFS7/NDUFV1 | 7 |
| MeSH_gene2pubmed | B_8_2 | D019427 | HIV Integrase | 5/48 | 109/26076 | 1.7265218188445e-06 | 0.00016666561027957 | 0.00014244705374516 | POLR2E/PSMA1/PSMB5/PSMC1/PSMD9 | 5 |
| GO | B_8_2 | GO:0006119 | oxidative phosphorylation | 6/47 | 149/18986 | 1.7391882055413e-06 | 6.22242891315888e-05 | 5.1361990865401e-05 | NDUFA11/NDUFA8/NDUFB9/NDUFS3/NDUFS7/NDUFV1 | 6 |
| MeSH_gene2pubmed | B_8_2 | D007968 | Leukoencephalopathy, Progressive Multifocal | 4/48 | 50/26076 | 2.18657506197691e-06 | 0.00016666561027957 | 0.00014244705374516 | DDX1/NDUFA8/NDUFS7/NDUFV1 | 4 |
| MeSH_gene2pubmed | B_8_2 | D016341 | Genes, vif | 4/48 | 50/26076 | 2.18657506197691e-06 | 0.00016666561027957 | 0.00014244705374516 | PSMA1/PSMB5/PSMC1/PSMD9 | 4 |
| GO | B_8_2 | GO:0009152 | purine ribonucleotide biosynthetic process | 6/47 | 156/18986 | 2.27168333493237e-06 | 7.55672979009732e-05 | 6.23757525988746e-05 | NDUFA11/NDUFA8/NDUFB9/NDUFS3/NDUFS7/NDUFV1 | 6 |
| KEGG | B_8_2 | hsa05022 | Pathways of neurodegeneration - multiple diseases | 10/29 | 483/8541 | 2.30283072495961e-06 | 2.07254765246365e-05 | 1.45441940523765e-05 | NDUFA11/NDUFA8/NDUFB9/NDUFS3/NDUFS7/NDUFV1/PSMA1/PSMB5/PSMC1/PSMD9 | 10 |