| reactome |
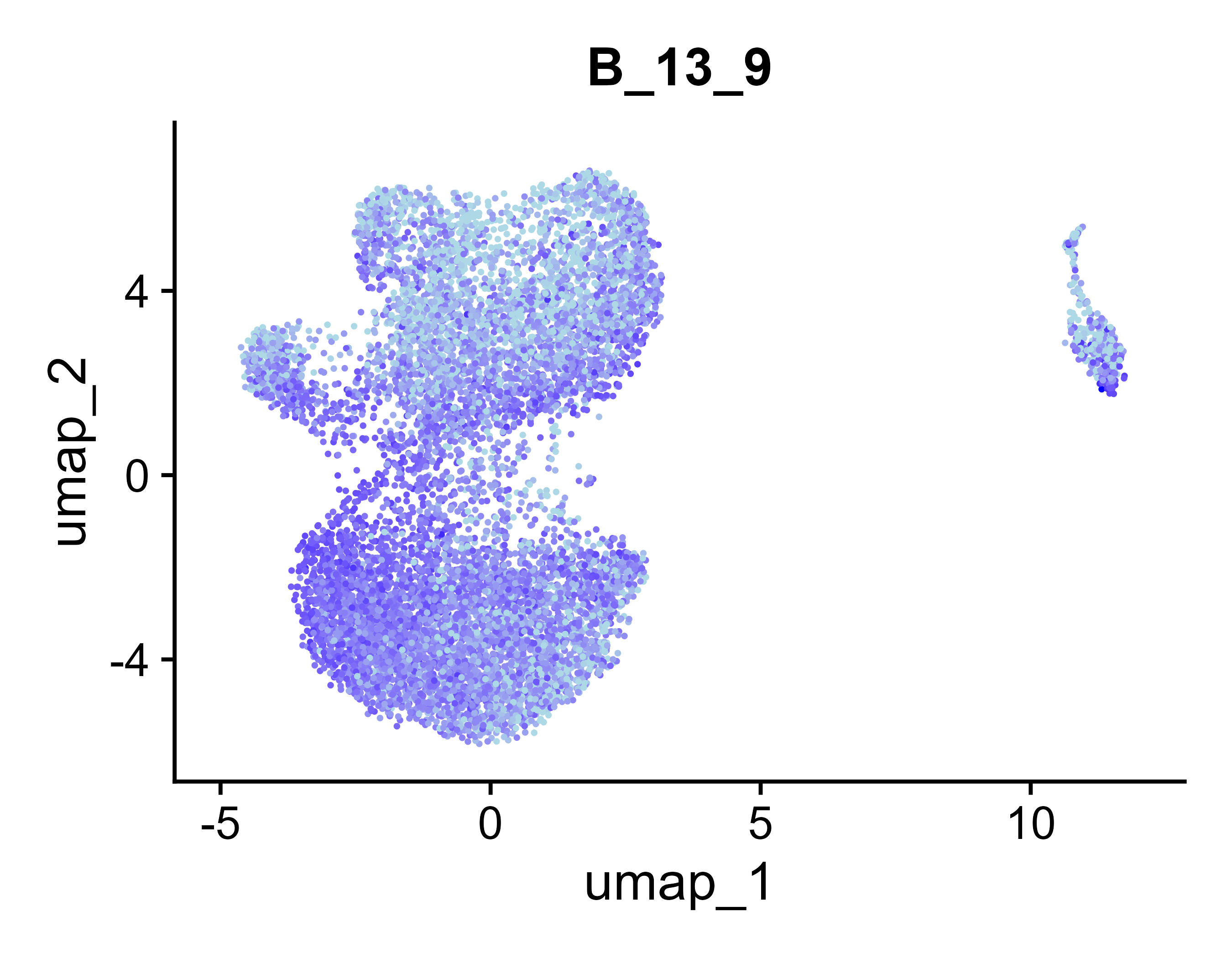
B_13_9 |
R-HSA-1226099 |
Signaling by FGFR in disease |
2/11 |
63/11146 |
0.00167351157214143 |
0.0362594173963976 |
0.0161479186785577 |
CPSF6/POLR2K |
2 |
| reactome |
B_13_9 |
R-HSA-5654738 |
Signaling by FGFR2 |
2/11 |
73/11146 |
0.00223981961046482 |
0.0380852860877859 |
0.0169610585816051 |
HNRNPM/POLR2K |
2 |
| reactome |
B_13_9 |
R-HSA-72203 |
Processing of Capped Intron-Containing Pre-mRNA |
3/11 |
285/11146 |
0.00234370991309452 |
0.0380852860877859 |
0.0169610585816051 |
CPSF6/HNRNPM/POLR2K |
3 |
| reactome |
B_13_9 |
R-HSA-9006931 |
Signaling by Nuclear Receptors |
3/11 |
298/11146 |
0.00266162737029145 |
0.0384457286819876 |
0.0171215795749742 |
DLD/POLR2K/PPP5C |
3 |
| reactome |
B_13_9 |
R-HSA-190236 |
Signaling by FGFR |
2/11 |
89/11146 |
0.00330893597895204 |
0.0430161677263765 |
0.0191569977728802 |
HNRNPM/POLR2K |
2 |
| KEGG |
B_13_9 |
hsa01200 |
Carbon metabolism |
2/8 |
116/8541 |
0.0048541364017801 |
0.0420691821487609 |
0.0238448805701479 |
DLD/IDH3B |
2 |
| hallmark |
B_13_9 |
HALLMARK_DNA_REPAIR |
HALLMARK_DNA_REPAIR |
2/5 |
150/4384 |
0.0108653944032605 |
0.0360439405264845 |
0.0189704950139392 |
POLR2K/TARBP2 |
2 |
| hallmark |
B_13_9 |
HALLMARK_FATTY_ACID_METABOLISM |
HALLMARK_FATTY_ACID_METABOLISM |
2/5 |
158/4384 |
0.0120146468421615 |
0.0360439405264845 |
0.0189704950139392 |
DLD/IDH3B |
2 |
| hallmark |
B_13_9 |
HALLMARK_OXIDATIVE_PHOSPHORYLATION |
HALLMARK_OXIDATIVE_PHOSPHORYLATION |
2/5 |
200/4384 |
0.0189034620878385 |
0.037806924175677 |
0.0198983811450931 |
DLD/IDH3B |
2 |