Module Explorer
Instructions
- Enter a module name.
- Click Submit to search for the detailed information of the module.
- Results will appear below if available.
Selected Module: B_12_22
Module Info
| Module_Name | B_12_22 |
| Cell_Type | B |
| Name | B_12_22: electron transfer activity |
| Meta_Module | B_3 |
| Gene_Number | 39 |
Genes:
ACTG1, ADH5, ANXA7, ARMC10, ATP6V0D1, BCL2L12, BOP1, C14orf119, CDC26, CHCHD3, CIAPIN1, CMAS, DERA, DHRS4L2, DMAC1, EIF2B1, EIF6, FERMT3, GRSF1, HMOX2, IARS2, LYPLA1, MRPS2, MYG1, NDUFA12, NT5DC1, PPT1, PSMD4, PSMG1, RBM42, RPP25L, SDHD, SMARCB1, TEX264, THOC7, TMEM126B, TMEM14A, WDR77, YIF1B
UMAP Visualizations

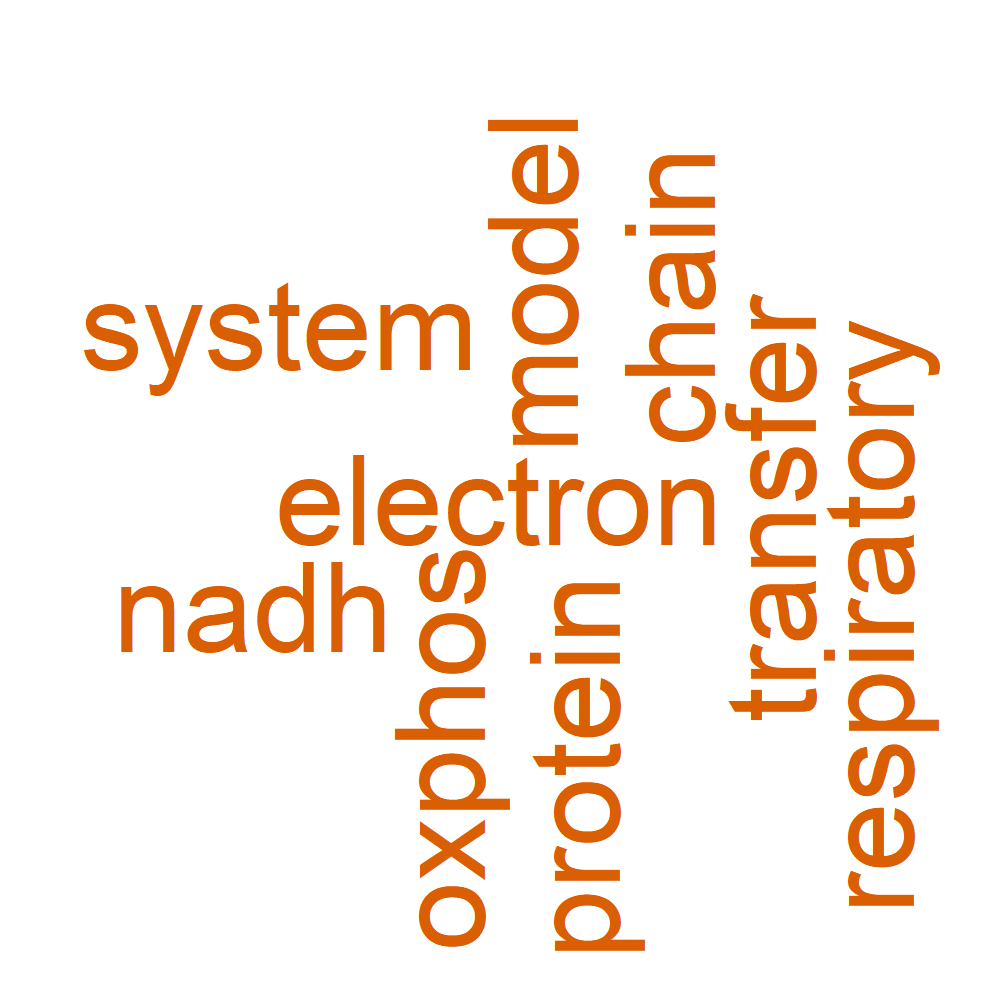
Word Cloud
Enriched TF
| TF | p.adjust | Genes |
|---|---|---|
| AP1 | 0.0408442135321342 | ANXA7/CMAS/FERMT3/PSMD4 |
| AP1 | 0.0408442135321342 | ANXA7/CMAS/FERMT3/PSMD4 |
Functional Enrichment Results
|
enrichment_term
|
module | ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count |
|---|---|---|---|---|---|---|---|---|---|---|
| GO | B_12_22 | GO:0009055 | electron transfer activity | 4/38 | 116/18737 | 8.75009635869114e-05 | 0.00962510599456026 | 0.00663165197711329 | ADH5/CIAPIN1/NDUFA12/SDHD | 4 |
| WikiPathways | B_12_22 | WP4324 | Mitochondrial complex I assembly model OXPHOS system | 3/17 | 56/8836 | 0.00015397435066525 | 0.00554307662394925 | 0.00437611312417046 | DMAC1/NDUFA12/TMEM126B | 3 |
| GO | B_12_22 | GO:0002084 | protein depalmitoylation | 2/37 | 10/18986 | 0.00016466503780535 | 0.0396352272928412 | 0.0308211345473018 | LYPLA1/PPT1 | 2 |
| GO | B_12_22 | GO:0042255 | ribosome assembly | 3/37 | 58/18986 | 0.00019526028398771 | 0.0396352272928412 | 0.0308211345473018 | BOP1/EIF6/MRPS2 | 3 |
| GO | B_12_22 | GO:0010257 | NADH dehydrogenase complex assembly | 3/37 | 62/18986 | 0.00023804941317021 | 0.0396352272928412 | 0.0308211345473018 | DMAC1/NDUFA12/TMEM126B | 3 |
| GO | B_12_22 | GO:0032981 | mitochondrial respiratory chain complex I assembly | 3/37 | 62/18986 | 0.00023804941317021 | 0.0396352272928412 | 0.0308211345473018 | DMAC1/NDUFA12/TMEM126B | 3 |
| GO | B_12_22 | GO:0098734 | macromolecule depalmitoylation | 2/37 | 14/18986 | 0.00033135697498618 | 0.04413674906816 | 0.0343216066722534 | LYPLA1/PPT1 | 2 |
| GO | B_12_22 | GO:0008474 | palmitoyl-(protein) hydrolase activity | 2/38 | 14/18737 | 0.00035890030449727 | 0.0131596778315666 | 0.00906695506098368 | LYPLA1/PPT1 | 2 |
| GO | B_12_22 | GO:0098599 | palmitoyl hydrolase activity | 2/38 | 14/18737 | 0.00035890030449727 | 0.0131596778315666 | 0.00906695506098368 | LYPLA1/PPT1 | 2 |
1