Module Explorer
Instructions
- Enter a module name.
- Click Submit to search for the detailed information of the module.
- Results will appear below if available.
Selected Module: B_12_13
Module Info
| Module_Name | B_12_13 |
| Cell_Type | B |
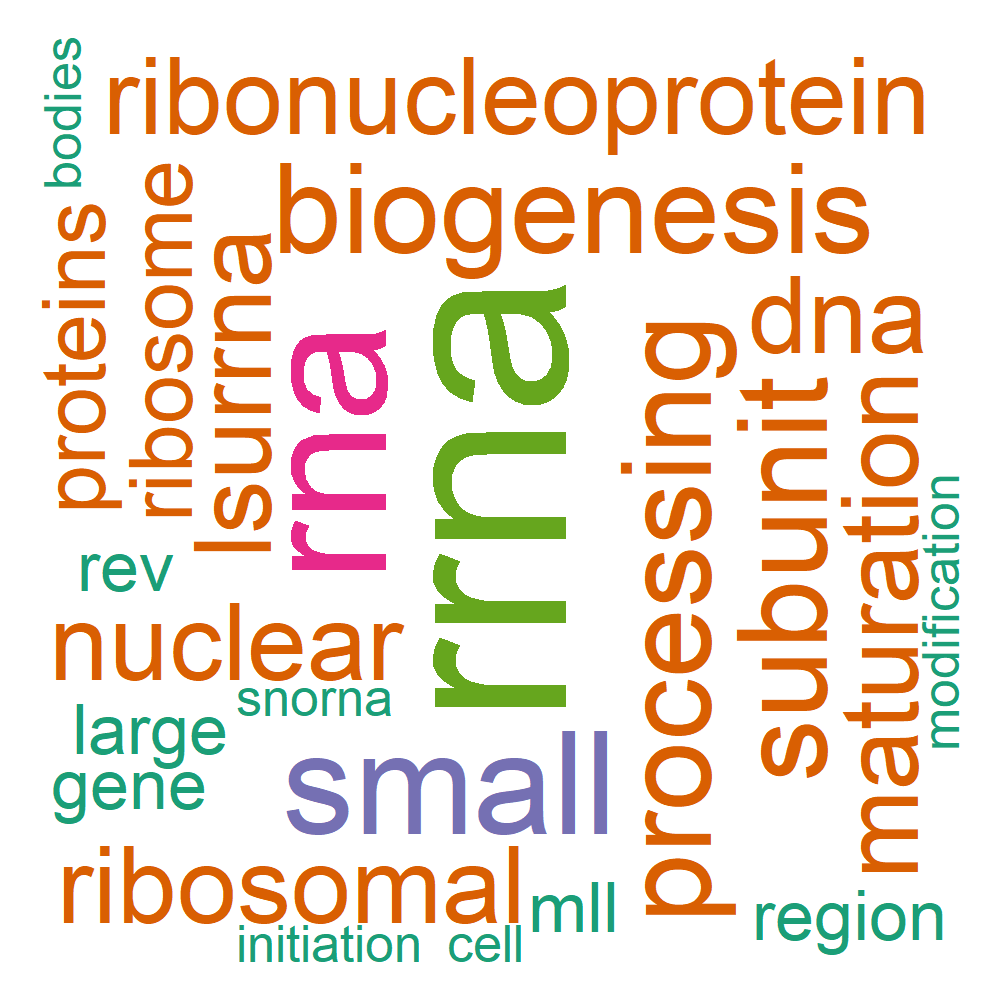
| Name | B_12_13: ribonucleoprotein complex biogenesis |
| Meta_Module | B_19 |
| Gene_Number | 60 |
Genes:
ANP32B, ATIC, AUP1, BANF1, CBX3, CDC123, CMSS1, COA4, COX20, DCTPP1, DDX56, DKC1, DPH2, EFTUD2, EIF3B, EXOSC7, FAM162A, FKBP4, GFM1, GPATCH4, HAT1, HDAC2, HDHD5, IFRD2, LAS1L, LTV1, LYAR, MFNG, MPHOSPH10, NCL, NDUFAF4, NIFK, NOC2L, NOLC1, NOP14, NOP16, NOP56, NPM1, NUDCD1, NUP62, ODC1, PAK1IP1, PES1, PNO1, PNPT1, POLD2, POLE3, PPP1R14B, RPF2, RPP30, RRP7A, RRP9, RUVBL1, SET, SNRPD3, SYNCRIP, TBRG4, TRIAP1, TRMT6, UMPS
UMAP Visualizations

Word Cloud
Enriched TF
| TF | p.adjust | Genes |
|---|---|---|
| MYCMAX | 2.5905944969521e-05 | DCTPP1/IFRD2/LYAR/NCL/NOP56/NPM1/ODC1/SYNCRIP |
| E2F1 | 4.15633532006135e-05 | CBX3/DCTPP1/NCL/NOLC1/NUP62/SYNCRIP/TRMT6 |
| E2F | 4.15633532006135e-05 | CBX3/DCTPP1/NCL/NOLC1/NUP62/SYNCRIP/TRMT6 |
| E2F1DP1 | 4.15633532006135e-05 | CBX3/DCTPP1/NCL/NOLC1/NUP62/SYNCRIP/TRMT6 |
| E2F1DP2 | 4.15633532006135e-05 | CBX3/DCTPP1/NCL/NOLC1/NUP62/SYNCRIP/TRMT6 |
| E2F4DP2 | 4.15633532006135e-05 | CBX3/DCTPP1/NCL/NOLC1/NUP62/SYNCRIP/TRMT6 |
| E2F4DP1 | 4.15633532006135e-05 | CBX3/DCTPP1/NCL/NOLC1/NUP62/SYNCRIP/TRMT6 |
| EPC1 | 0.00084259806510092 | ANP32B/BANF1/CMSS1/HDAC2/NCL/NOP56/TRIAP1 |
Functional Enrichment Results
|
enrichment_term
|
module | ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count |
|---|---|---|---|---|---|---|---|---|---|---|
| MeSH_gene2pubmed | B_12_13 | D009697 | Nucleolus Organizer Region | 4/60 | 61/26076 | 1.19802823687656e-05 | 0.00142685163011999 | 0.00124973261341545 | NCL/NOLC1/NPM1/RRP9 | 4 |
| hallmark | B_12_13 | HALLMARK_UNFOLDED_PROTEIN_RESPONSE | HALLMARK_UNFOLDED_PROTEIN_RESPONSE | 7/32 | 113/4384 | 1.2394204884079e-05 | 7.84966309325001e-05 | 6.0883813465651e-05 | BANF1/DKC1/NOLC1/NOP14/NOP56/NPM1/RRP9 | 7 |
| GO | B_12_13 | GO:0042393 | histone binding | 7/59 | 249/18737 | 1.27390472968281e-05 | 0.00107007997293356 | 0.00075763807607451 | ANP32B/CBX3/HAT1/HDAC2/NOC2L/NPM1/SET | 7 |
| GO | B_12_13 | GO:0034708 | methyltransferase complex | 5/59 | 103/19960 | 1.33153922750566e-05 | 0.0003302217284214 | 0.00016819442873755 | HDAC2/LAS1L/RUVBL1/SNRPD3/TRMT6 | 5 |
| GO | B_12_13 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 3/58 | 17/18986 | 1.78463919187796e-05 | 0.00155977465370133 | 0.00126615454244815 | PAK1IP1/PES1/RPF2 | 3 |
| MeSH_gene2pubmed | B_12_13 | D020035 | Holoenzymes | 5/60 | 143/26076 | 1.98125177065872e-05 | 0.00214515532623139 | 0.00187887129638544 | DKC1/NCL/PPP1R14B/RPP30/RUVBL1 | 5 |
| MeSH_gene2pubmed | B_12_13 | D012318 | RNA Polymerase I | 5/60 | 151/26076 | 2.5744835182747e-05 | 0.00255517489188764 | 0.00223799400579845 | NCL/NOLC1/RRP9/RUVBL1/SET | 5 |
| MeSH_gene2pubmed | B_12_13 | D012337 | RNA, Ribosomal, 18S | 5/60 | 155/26076 | 2.91858932221804e-05 | 0.00267387683289361 | 0.00234196114843569 | DDX56/MPHOSPH10/NCL/NOP14/RRP9 | 5 |
| GO | B_12_13 | GO:0006334 | nucleosome assembly | 5/58 | 120/18986 | 3.25226508430927e-05 | 0.00258407243971482 | 0.00209763317399469 | ANP32B/HAT1/NPM1/POLE3/SET | 5 |
| MeSH_gene2pubmed | B_12_13 | D017381 | Spliceosomes | 7/60 | 422/26076 | 5.10229704670163e-05 | 0.00407276096294241 | 0.00356719794447473 | CBX3/EFTUD2/HDAC2/NPM1/RRP9/SNRPD3/SYNCRIP | 7 |