Module Explorer
Instructions
- Enter a module name.
- Click Submit to search for the detailed information of the module.
- Results will appear below if available.
Selected Module: B_12_13
Module Info
| Module_Name | B_12_13 |
| Cell_Type | B |
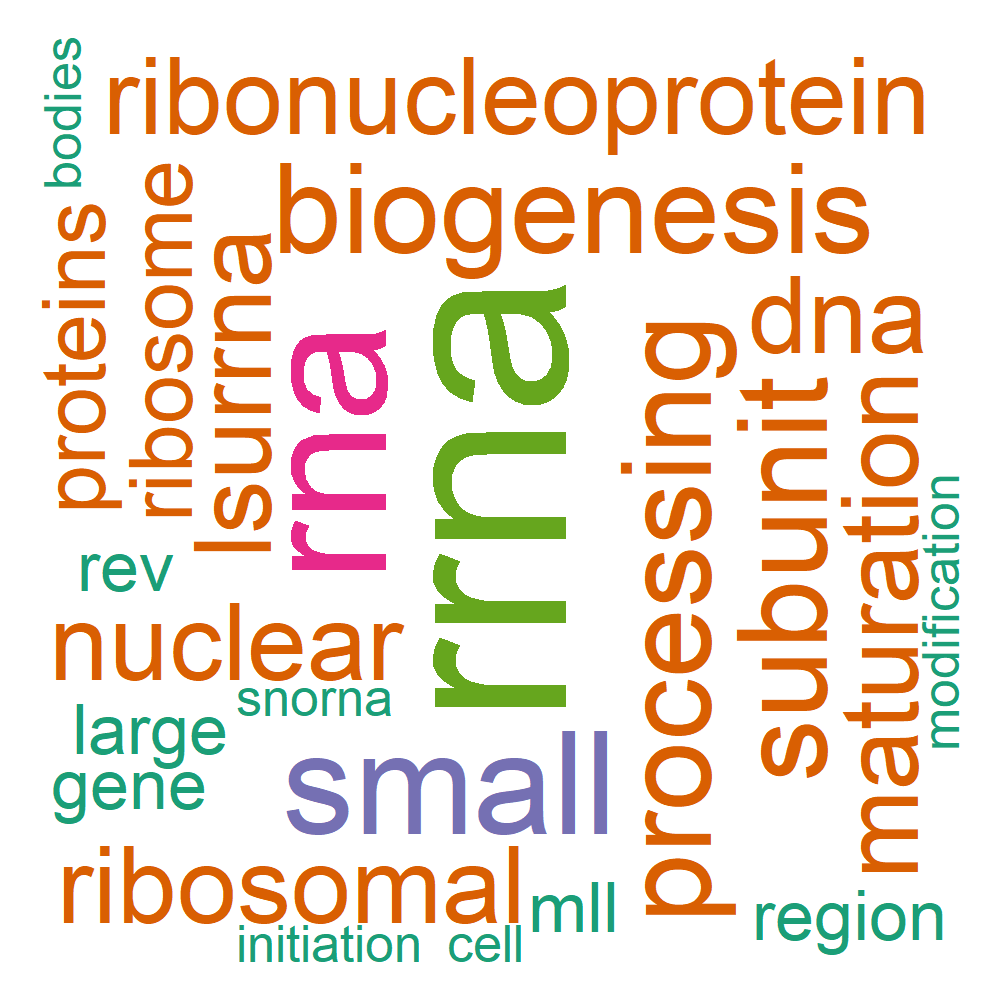
| Name | B_12_13: ribonucleoprotein complex biogenesis |
| Meta_Module | B_19 |
| Gene_Number | 60 |
Genes:
ANP32B, ATIC, AUP1, BANF1, CBX3, CDC123, CMSS1, COA4, COX20, DCTPP1, DDX56, DKC1, DPH2, EFTUD2, EIF3B, EXOSC7, FAM162A, FKBP4, GFM1, GPATCH4, HAT1, HDAC2, HDHD5, IFRD2, LAS1L, LTV1, LYAR, MFNG, MPHOSPH10, NCL, NDUFAF4, NIFK, NOC2L, NOLC1, NOP14, NOP16, NOP56, NPM1, NUDCD1, NUP62, ODC1, PAK1IP1, PES1, PNO1, PNPT1, POLD2, POLE3, PPP1R14B, RPF2, RPP30, RRP7A, RRP9, RUVBL1, SET, SNRPD3, SYNCRIP, TBRG4, TRIAP1, TRMT6, UMPS
UMAP Visualizations

Word Cloud
Enriched TF
| TF | p.adjust | Genes |
|---|---|---|
| MYCMAX | 2.5905944969521e-05 | DCTPP1/IFRD2/LYAR/NCL/NOP56/NPM1/ODC1/SYNCRIP |
| E2F1 | 4.15633532006135e-05 | CBX3/DCTPP1/NCL/NOLC1/NUP62/SYNCRIP/TRMT6 |
| E2F | 4.15633532006135e-05 | CBX3/DCTPP1/NCL/NOLC1/NUP62/SYNCRIP/TRMT6 |
| E2F1DP1 | 4.15633532006135e-05 | CBX3/DCTPP1/NCL/NOLC1/NUP62/SYNCRIP/TRMT6 |
| E2F1DP2 | 4.15633532006135e-05 | CBX3/DCTPP1/NCL/NOLC1/NUP62/SYNCRIP/TRMT6 |
| E2F4DP2 | 4.15633532006135e-05 | CBX3/DCTPP1/NCL/NOLC1/NUP62/SYNCRIP/TRMT6 |
| E2F4DP1 | 4.15633532006135e-05 | CBX3/DCTPP1/NCL/NOLC1/NUP62/SYNCRIP/TRMT6 |
| EPC1 | 0.00084259806510092 | ANP32B/BANF1/CMSS1/HDAC2/NCL/NOP56/TRIAP1 |
Functional Enrichment Results
|
enrichment_term
|
module | ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count |
|---|---|---|---|---|---|---|---|---|---|---|
| MeSH_gene2pubmed | B_12_13 | D034441 | Heterogeneous-Nuclear Ribonucleoproteins | 9/60 | 450/26076 | 8.4996782317248e-07 | 0.00020246233547968 | 0.00017733012900293 | EFTUD2/NCL/NOLC1/NPM1/NUP62/RRP9/SET/SNRPD3/SYNCRIP | 9 |
| MeSH_gene2pubmed | B_12_13 | D053487 | DEAD-box RNA Helicases | 9/60 | 465/26076 | 1.11465274383182e-06 | 0.00022049471178885 | 0.00019312409685161 | DDX56/EFTUD2/HDAC2/NCL/NPM1/NUP62/ODC1/RRP9/SNRPD3 | 9 |
| GO | B_12_13 | GO:0000470 | maturation of LSU-rRNA | 4/58 | 26/18986 | 1.11465550532809e-06 | 0.00013917270166525 | 0.00011297410685581 | LAS1L/PAK1IP1/PES1/RPF2 | 4 |
| MeSH_gene2pubmed | B_12_13 | D035463 | Pol1 Transcription Initiation Complex Proteins | 5/60 | 82/26076 | 1.29593869229385e-06 | 0.00022049471178885 | 0.00019312409685161 | NCL/NOLC1/RRP9/RUVBL1/SET | 5 |
| GO | B_12_13 | GO:0030515 | snoRNA binding | 4/59 | 32/18737 | 2.98487979716818e-06 | 0.00050145980592425 | 0.00035504359692632 | DKC1/NOP14/NOP56/RRP9 | 4 |
| GO | B_12_13 | GO:0000460 | maturation of 5.8S rRNA | 4/58 | 35/18986 | 3.82477205383131e-06 | 0.00041785634688107 | 0.00033919689003714 | EXOSC7/LAS1L/NOP14/PES1 | 4 |
| MeSH_gene2pubmed | B_12_13 | D012331 | RNA, Fungal | 6/60 | 188/26076 | 4.69764857508113e-06 | 0.0006993624316152 | 0.00061254864972439 | CBX3/DPH2/NOP14/PES1/RPP30/RRP9 | 6 |
| GO | B_12_13 | GO:0000966 | RNA 5'-end processing | 4/58 | 38/18986 | 5.35533903746073e-06 | 0.00052006292430451 | 0.00042216356856707 | NOP14/PNPT1/RPP30/SNRPD3 | 4 |
| WikiPathways | B_12_13 | WP4022 | Pyrimidine metabolism | 5/29 | 84/8836 | 6.83481753246438e-06 | 0.00041692386948032 | 0.00032375451469568 | DCTPP1/PNPT1/POLD2/POLE3/UMPS | 5 |
| MeSH_gene2pubmed | B_12_13 | D036961 | Matrix Attachment Region Binding Proteins | 5/60 | 121/26076 | 8.81670024718132e-06 | 0.00116674333271033 | 0.00102191227426394 | EFTUD2/HDAC2/NCL/NPM1/SNRPD3 | 5 |