Module Explorer
Instructions
- Enter a module name.
- Click Submit to search for the detailed information of the module.
- Results will appear below if available.
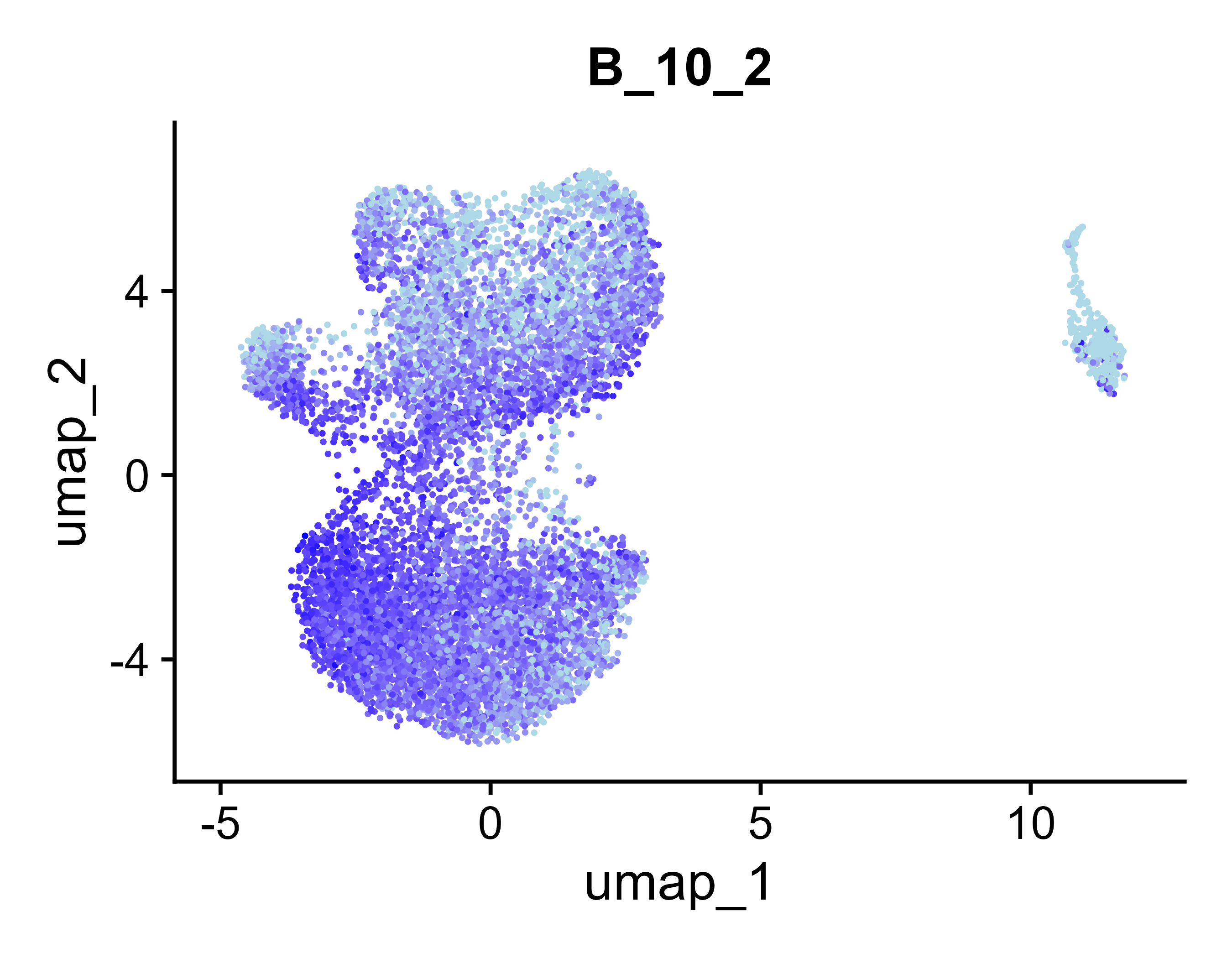
Selected Module: B_10_2
Module Info
| Module_Name | B_10_2 |
| Cell_Type | B |
| Name | B_10_2: mitochondrial protein-containing complex |
| Meta_Module | B_3 |
| Gene_Number | 76 |
Genes:
ACAA2, AHSA1, APIP, ARPC5L, ATP5F1A, BCAP31, C16orf87, CACYBP, CCDC25, CHCHD10, CLTA, CMC2, COPS6, DCTN2, DDX54, ELAVL1, ELP6, EMC8, FKBP3, GLRX3, GPS1, HDDC2, HDDC3, HIKESHI, HPF1, HSPA8, IMMP1L, IMPDH1, KXD1, LSM12, METTL26, METTL5, MRPL22, MRPL34, MRPL39, MRPL43, MRPL47, MRPS10, MRPS18B, MRPS18C, MRPS5, MRPS6, MTX2, NAA20, NDUFA10, NDUFB5, NDUFS4, NELFE, NUP37, ORMDL2, PDHB, POP4, PPA2, PSMC5, PSMD13, RAB8A, RBX1, RNF5, RWDD1, SAMM50, SKIC8, SNAP29, SNRPB2, STIP1, SUGT1, TAF12, TMA16, TMX1, TOR1A, TPRKB, TRMT112, TTC1, TXNDC12, UBE2V2, UQCC3, VCF1
UMAP Visualizations

Word Cloud
Enriched TF
| TF | p.adjust | Genes |
|---|---|---|
| PPARGC1A | 0.00014596863321504 | AHSA1/CACYBP/HIKESHI/HSPA8/MRPS6/STIP1 |
| FOXR2 | 0.0085165337658232 | AHSA1/MRPL43/NELFE/PSMC5/TAF12/TRMT112 |
Functional Enrichment Results
|
enrichment_term
|
module | ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count |
|---|---|---|---|---|---|---|---|---|---|---|
| GO | B_10_2 | GO:0098799 | outer mitochondrial membrane protein complex | 2/71 | 23/19960 | 0.0030073636171011 | 0.0290711816319773 | 0.0228190309543198 | MTX2/SAMM50 | 2 |
| reactome | B_10_2 | R-HSA-5610780 | Degradation of GLI1 by the proteasome | 3/54 | 60/11146 | 0.00302677423598843 | 0.0402560973386462 | 0.0299491345455697 | PSMC5/PSMD13/RBX1 | 3 |
| reactome | B_10_2 | R-HSA-5610783 | Degradation of GLI2 by the proteasome | 3/54 | 60/11146 | 0.00302677423598843 | 0.0402560973386462 | 0.0299491345455697 | PSMC5/PSMD13/RBX1 | 3 |
| reactome | B_10_2 | R-HSA-5610785 | GLI3 is processed to GLI3R by the proteasome | 3/54 | 60/11146 | 0.00302677423598843 | 0.0402560973386462 | 0.0299491345455697 | PSMC5/PSMD13/RBX1 | 3 |
| GO | B_10_2 | GO:0034976 | response to endoplasmic reticulum stress | 5/68 | 277/18986 | 0.0031427502437425 | 0.0444040195728779 | 0.0380438187400408 | BCAP31/RNF5/TMX1/TOR1A/TXNDC12 | 5 |
| reactome | B_10_2 | R-HSA-5678895 | Defective CFTR causes cystic fibrosis | 3/54 | 61/11146 | 0.00317250697952131 | 0.0408332349944839 | 0.030378504863328 | PSMC5/PSMD13/RNF5 | 3 |
| GO | B_10_2 | GO:1901028 | regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 2/68 | 24/18986 | 0.00331540797315846 | 0.0460999584839176 | 0.0394968401814867 | ACAA2/CHCHD10 | 2 |
| KEGG | B_10_2 | hsa05022 | Pathways of neurodegeneration - multiple diseases | 8/45 | 483/8541 | 0.00333449695249093 | 0.0395376067223925 | 0.0330942554683311 | ATP5F1A/DCTN2/NDUFA10/NDUFB5/NDUFS4/PSMC5/PSMD13/RAB8A | 8 |
| GO | B_10_2 | GO:0009451 | RNA modification | 4/68 | 173/18986 | 0.00344544597539941 | 0.0471595417882795 | 0.040404654283878 | ELP6/METTL5/TPRKB/TRMT112 | 4 |
| GO | B_10_2 | GO:0022624 | proteasome accessory complex | 2/71 | 25/19960 | 0.00354971368121058 | 0.0329413429616342 | 0.0258568617620813 | PSMC5/PSMD13 | 2 |