Module Explorer
Instructions
- Enter a module name.
- Click Submit to search for the detailed information of the module.
- Results will appear below if available.
Selected Module: B_10_2
Module Info
| Module_Name | B_10_2 |
| Cell_Type | B |
| Name | B_10_2: mitochondrial protein-containing complex |
| Meta_Module | B_3 |
| Gene_Number | 76 |
Genes:
ACAA2, AHSA1, APIP, ARPC5L, ATP5F1A, BCAP31, C16orf87, CACYBP, CCDC25, CHCHD10, CLTA, CMC2, COPS6, DCTN2, DDX54, ELAVL1, ELP6, EMC8, FKBP3, GLRX3, GPS1, HDDC2, HDDC3, HIKESHI, HPF1, HSPA8, IMMP1L, IMPDH1, KXD1, LSM12, METTL26, METTL5, MRPL22, MRPL34, MRPL39, MRPL43, MRPL47, MRPS10, MRPS18B, MRPS18C, MRPS5, MRPS6, MTX2, NAA20, NDUFA10, NDUFB5, NDUFS4, NELFE, NUP37, ORMDL2, PDHB, POP4, PPA2, PSMC5, PSMD13, RAB8A, RBX1, RNF5, RWDD1, SAMM50, SKIC8, SNAP29, SNRPB2, STIP1, SUGT1, TAF12, TMA16, TMX1, TOR1A, TPRKB, TRMT112, TTC1, TXNDC12, UBE2V2, UQCC3, VCF1
UMAP Visualizations

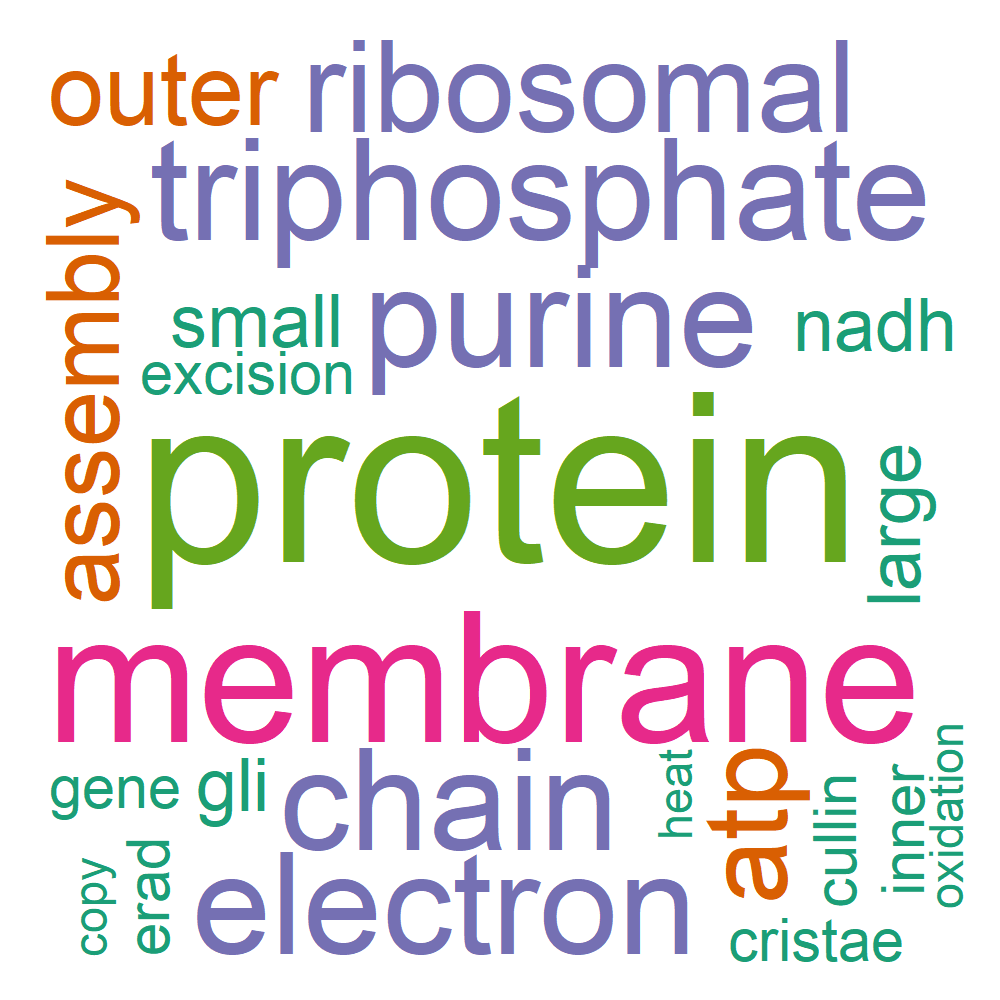
Word Cloud
Enriched TF
| TF | p.adjust | Genes |
|---|---|---|
| PPARGC1A | 0.00014596863321504 | AHSA1/CACYBP/HIKESHI/HSPA8/MRPS6/STIP1 |
| FOXR2 | 0.0085165337658232 | AHSA1/MRPL43/NELFE/PSMC5/TAF12/TRMT112 |
Functional Enrichment Results
|
enrichment_term
|
module | ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count |
|---|---|---|---|---|---|---|---|---|---|---|
| reactome | B_10_2 | R-HSA-6781823 | Formation of TC-NER Pre-Incision Complex | 3/54 | 52/11146 | 0.00200879582132246 | 0.0396647647367806 | 0.0295092036822902 | COPS6/GPS1/RBX1 | 3 |
| reactome | B_10_2 | R-HSA-9762114 | GSK3B and BTRC:CUL1-mediated-degradation of NFE2L2 | 3/54 | 52/11146 | 0.00200879582132246 | 0.0396647647367806 | 0.0295092036822902 | PSMC5/PSMD13/RBX1 | 3 |
| GO | B_10_2 | GO:1990204 | oxidoreductase complex | 4/71 | 150/19960 | 0.00201482771870413 | 0.0233720015369679 | 0.0183455365966218 | NDUFA10/NDUFB5/NDUFS4/PDHB | 4 |
| GO | B_10_2 | GO:0009117 | nucleotide metabolic process | 7/68 | 500/18986 | 0.00204133959322269 | 0.0308312669597082 | 0.0264151566237346 | ATP5F1A/HSPA8/IMPDH1/NDUFA10/NDUFB5/NDUFS4/UQCC3 | 7 |
| GO | B_10_2 | GO:1904294 | positive regulation of ERAD pathway | 2/68 | 19/18986 | 0.00207796652343469 | 0.0308525199072676 | 0.0264333654096152 | BCAP31/TMX1 | 2 |
| WikiPathways | B_10_2 | WP5381 | Smith Magenis and Potocki Lupski syndrome copy number variation | 4/31 | 159/8836 | 0.00217521794578324 | 0.0253775427008044 | 0.0221337966413031 | CLTA/COPS6/GPS1/RAB8A | 4 |
| reactome | B_10_2 | R-HSA-180585 | Vif-mediated degradation of APOBEC3G | 3/54 | 54/11146 | 0.00223925131368399 | 0.0396647647367806 | 0.0295092036822902 | PSMC5/PSMD13/RBX1 | 3 |
| reactome | B_10_2 | R-HSA-9604323 | Negative regulation of NOTCH4 signaling | 3/54 | 54/11146 | 0.00223925131368399 | 0.0396647647367806 | 0.0295092036822902 | PSMC5/PSMD13/RBX1 | 3 |
| GO | B_10_2 | GO:2000434 | regulation of protein neddylation | 2/68 | 20/18986 | 0.00230352333470246 | 0.0336314406866559 | 0.028814248028822 | COPS6/GPS1 | 2 |
| reactome | B_10_2 | R-HSA-3371497 | HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand | 3/54 | 55/11146 | 0.00236037693393242 | 0.0396647647367806 | 0.0295092036822902 | DCTN2/HSPA8/STIP1 | 3 |